Fig. 5. Sperm miRNAs from depression model cause a transcriptional cascade change during early embryonic development.
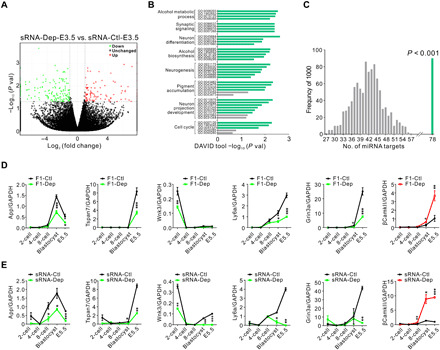
(A) Scatter plot comparison illustrating differentially expressed genes in sRNA-Dep-E3.5 versus sRNA-Ctl-E3.5. Down-regulated genes are shown in green, and up-regulated genes are shown in red. (B) Functional annotation clustering of the differentially expressed genes according to GO categories. The x axis represents the negative log of the P values of the enrichment of the corresponding GO terms. The eight top-ranked GO clusters are listed. Green bars indicate P < 0.05; gray bars indicate P > 0.05 for unique GO terms. (C) Number of 264 random genes (107 up-regulated and 157 down-regulated) targeted by the 17-miRNA set (16 up-regulated and 1 down-regulated). Genes potentially targeted by the 17-miRNA set were predicted using TargetScan, and the output is compared to genuinely changed or randomly selected gene set for overlapping genes. The simulation results of 1000 replicates are shown in gray, and the observed genuine value is in green. (D and E) Line chart of the relative expression levels of App, Tspan7, Wnk3, Ly6a, Grin3a, and βCamkII in early embryos. Zygotes derived from F0-Dep or F0-Ctl (D) or injected with sperm sRNA from F0-Dep or F0-Ctl (E) were cultured, and the gene transcriptional changes were assessed across early embryonic stages (n = 3 pools). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
