Fig. 3. ESMD1 is a positive regulator of ectopic JA-Ile production in kor1.
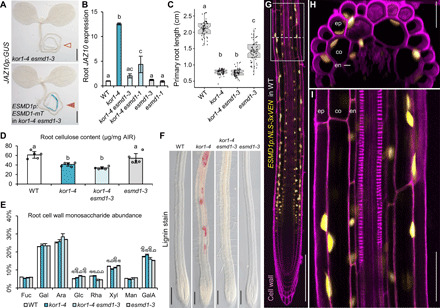
(A) JAZ10p:GUS expression is absent in kor1-4 esmd1-3 (empty arrowhead) and present in its ESMD1p:ESMD1-mTurquoise (mT) complementation line (orange arrowhead). (B) qRT-PCR of basal JAZ10 expression in roots of indicated genotypes. JAZ10 transcript levels were normalized to UBC21. Bars represent the means of three biological replicates (±SD), each containing ~60 roots from 5-do seedlings. (C) Primary root length box plots from 7-do seedlings of indicated genotypes. Medians are represented inside the boxes by solid lines, and circles depict individual measurements (n = 58 to 66). (D) Crystalline cellulose content from alcohol-insoluble residue (AIR) extracted from roots of indicated genotypes. Bars represent means of five biological replicates depicted as circles (±SD), each consisting of pools from ~300 roots from 12-do seedlings. (E) Cell wall monosaccharide composition analysis from root-extracted AIR of indicated genotypes. Bars represent the means of three biological replicates (±SD). Only statistically significant differences among genotypes indicated with letters and assessed for each individual sugar are shown above bars. Fuc, fucose; Gal, galactose; Ara, arabinose; Glc, glucose; Rha, rhamnose; Xyl, xylose; Man, mannose; GalA, galacturonic acid. (F) Lignin deposition visualized by phloroglucinol stain (fuchsia) in primary roots of indicated genotypes. (G to I) ESMD1p:NLS-3xVEN expression in 5-do WT primary roots counterstained with PI. (H) Orthogonal view from epidermis to vascular cylinder of a Z-stack section in the early differentiation zone through (G) (dotted line). (I) Increased magnification from (G) (boxed). ep, epidermis; co, cortex; en, endodermis. Letters in (B) to (E) denote statistically significant differences among samples as determined by ANOVA followed by Tukey’s HSD test [P < 0.05 and P < 0.001 in (E)]. Scale bars, 0.5 mm (A), 200 μm (F and G), 25 μm (H), and 50 μm (I).
