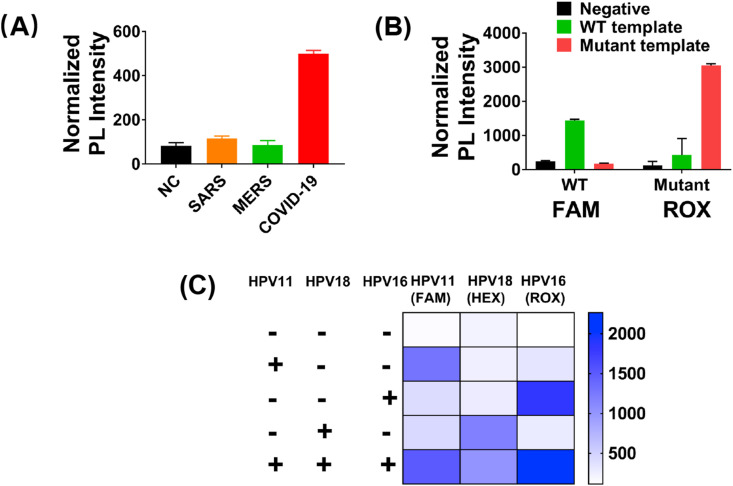
Fig. 4.
Analysis of the specificity of PLCR. (A) The ability of PLCR to distinguish SARS-CoV-2 and other coronaviruses. The nucleic acid concentrations of SARS-CoV-2, MERS, and SARS used in the experiment were 1 pM. The LCR was carried out at 80 °C 5 s, 40 °C 1 min for 30 thermal cycles, followed with PfAgo digestion for 20 min. (B) The ability of PLCR to distinguish wild type SARS-CoV-2 and its spike D614G mutant. The nucleic acid concentrations of SARS-CoV-2 and its mutant used in the experiment were 1 pM. The LCR was carried out at 80 °C 5 s, 40 °C 1 min for 30 thermal cycles, followed with PfAgo digestion for 40 min. (C) The results of PLCR with three-channel detection about HPV different subtypes. The nucleic acid concentrations of HPV11, HPV16 and HPV18 used in the experiment were 1 pM. The LCR was carried out at 80 °C 5 s, 40 °C 1 min for 30 thermal cycles, followed with PfAgo digestion for 40 min. Data are presented as mean ± SD of three technical replicates.