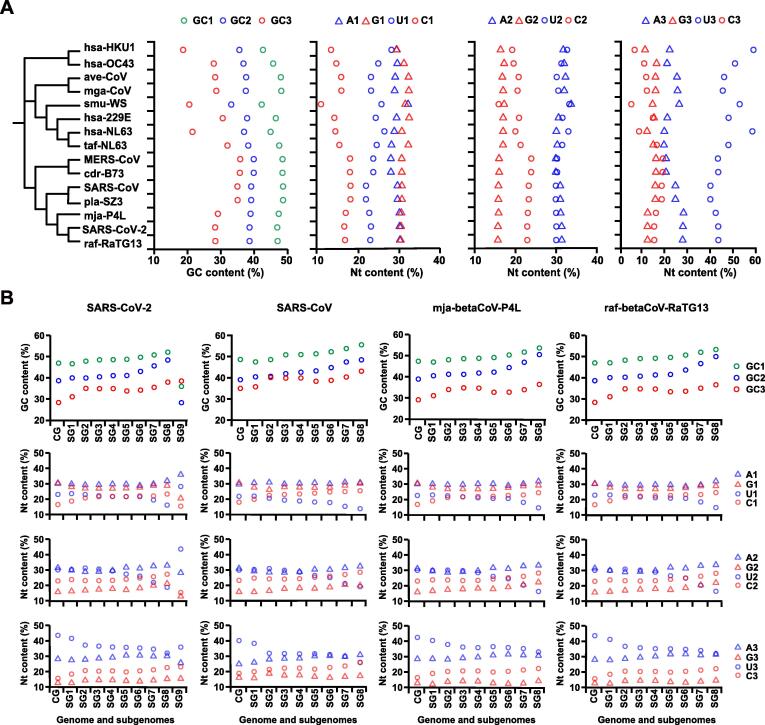
Figure 2.
Nucleotide contents of genomes and subgenomes of SARS-CoV-2 and related CoVs
A. A schematic phylogenetic tree is used to cluster genome sequences and compositional variables (15 CoVs genome sequences, from top to bottom, are: hsa-betaCoV-HKU1, hsa-betaCoV-OC43, ave-gamaCoV, mga-gamaCoV, smu-alphaCoV-WS, hsa-alphaCoV-229E, hsa-alphaCoV-NL63, taf-alphaCoV-NL63, MERS-CoV, cdr-betaCoV-B73, SARS-CoV, pla-betaCoV-SZ3, mja-betaCoV-P4L, SARS-CoV-2, and raf-betaCoV-RaTG13). These compositional variables include GC1, GC2, and GC3 and single nucleotide contents at three codon positions (A1, A2, A3; U1, U2, U3; G1, G2, G3; and C1, C2, C3). Nucleotides are labeled in different shapes: purines, triangles; pyrimidines, open circles. A and U or G and C are colored blue or red, respectively. It becomes obvious that the two closely-related CoV genomes to SARS-CoV-2, the reported bat (raf-betaCoV-RaTG13) and the pangolin (mja-betaCoV-P4L), have very similar codon G + C contents as well as base contents. The CP1 (codon position 1) base content appears most characteristic of balanced purine content of SARS-CoV-2 and its close relatives. The CP2 (codon position 2) base content of SARS-CoV-2 and all other CoVs has higher and relatively balanced A + U content. The older human CoVs have either lowest or higher G + C content and unbalanced purine content. G + C content represents a single measure but single nucleotide content demonstrates trends of all four nucleotides. B. The G + C and single nucleotide contents at different codon positions of complete genomes (labeled as “CG”) and subgenomes (labeled as “SG”) of SARS-CoV-2, SARS-CoV, mja-betaCoV-P4L, and raf-betaCoV-RaTG13 are displayed to illustrate the driving force for G + C content decrease towards 3’ end of the genome, which is rather a result of, in terms of mechanism, the increased U content and C-to-U permutation. The negative gradient of U is also obvious from the 5′ end to the 3′ end.