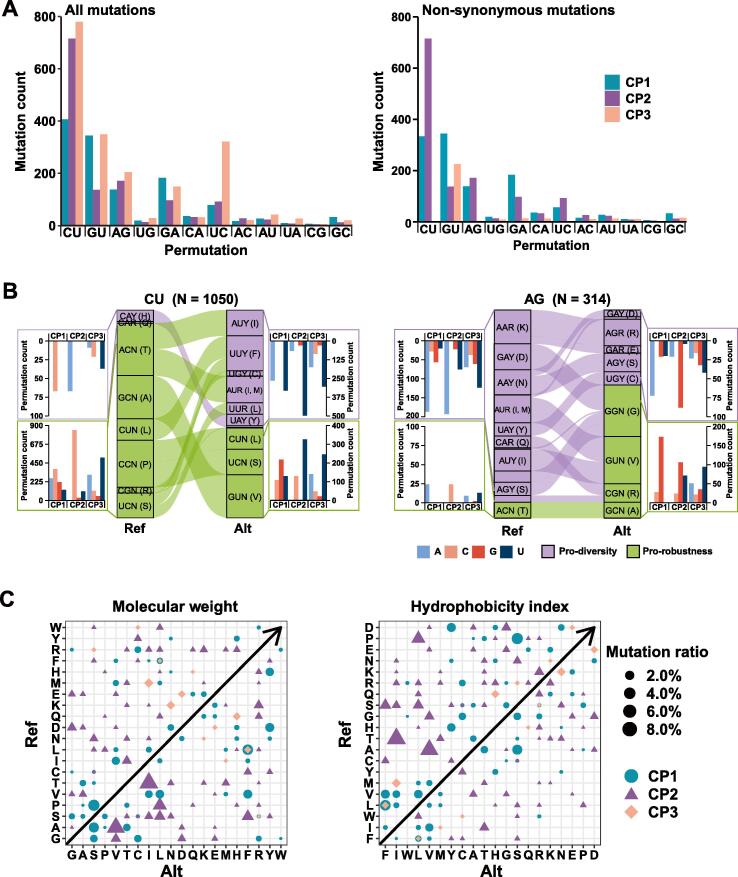
Figure 3.
Mutation spectra of SARS-CoV-2 in a context of codon positions
A. The SARS-CoV-2 mutation spectrum is composed of 12 permutations and they are divided by codon positions among all mutations. For all mutations, C-to-U (CU), U-to-C (UC), A-to-G (AG), G-to-A (GA), and G-to-U (GU), are always dominant due to two principles; one is that the first four permutations occur when positive-sense genome is synthesized, and the other is that a G-by-A replacement is always preferred by RTCs so that G-to-U permutation is the most dominant when the antigenome serves as a template. For non-synonymous mutations, C-to-U permutations at CP3 diminish among non-synonymous mutations and this phenomenon indicates that most protein composition relevant variations are CP1 and CP2 variations. The remaining non-synonymous mutations in G-to-U (GU) permutation may be a result of biased strand synthesis. B. Displays of permutation-to-codon changes among non-synonymous mutations. The codon table is divided into two halves: the pro-diversity half (purple) whose CP3 is sensitive to transitional change and the pro-robust half (green) whose CP3 position is insensitive to any change. Two examples, C-to-U (1051 in counts) and A-to-G (314 in counts) permutations are shown here. When a codon has a C-to-U change, the codon position varies, results of such changes relative to codon positions are summarized on both sides of the codon flow chart. Note that CP1 and CP2 changes appear more than those of CP3. C. All permutations are plotted against the reference genome sequence to show how changes are related to amino acids. In the molecular weight index, most CP1 and CP2 changes are showing an obvious increasing trend. In the hydrophobicity index, most CP1 and CP2 changes toward less hydrophobicity.