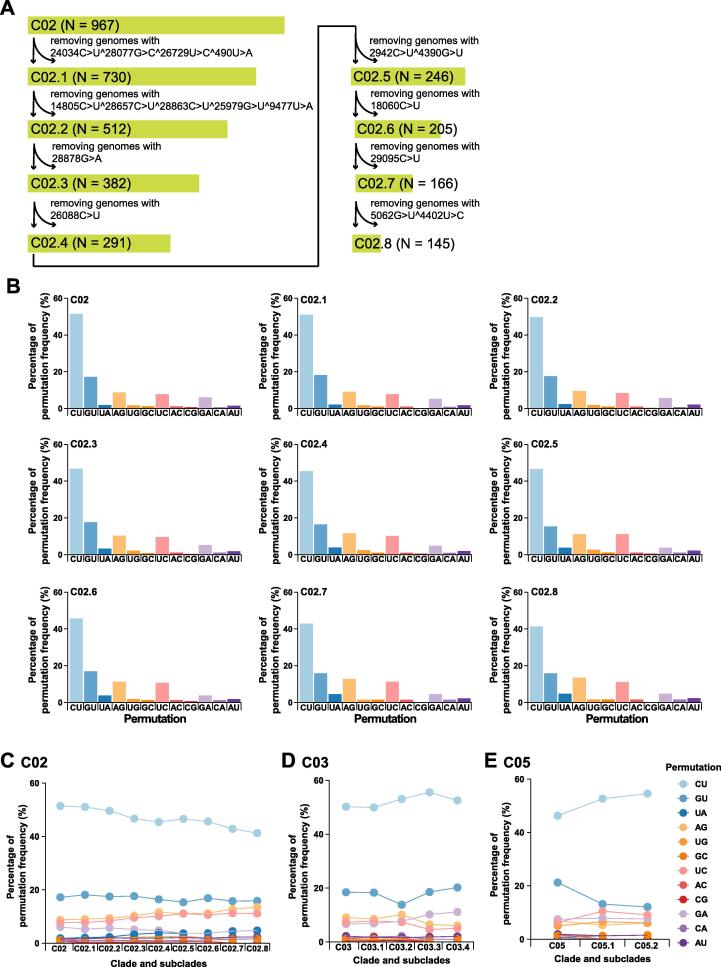
Figure 6.
Within-clade permutation variations are excellent indicators of functional mutations
A. The identification pipeline of subclades in clade C02. The number of SARS-CoV-2 genomes in each clade and subclade is indicated in the parentheses. B. Permutation variation of each subclade in clade C02. The clear trends are two-fold. First, decreased C-to-U permutation is coupled with increased A-to-G and decreased G-to-U permutations. Second, A-to-U permutation is also increased as expected based on the model shown in Figure S5. These trends of permutation changes suggest irrelevant to the ratio of strand-biased synthesis (positive sense vs negative sense) but possible structural and/ or conformational variations in the RTCs. C.–E. show within-clade permutation changes of C02, C03, and C05. In each display, the first column of the x-axis shows the proportion of permutations calculated for each clade. Two opposite trends of permutation variations are seen between C03 and C05, which has a rather wavy pattern.