FIGURE 3.
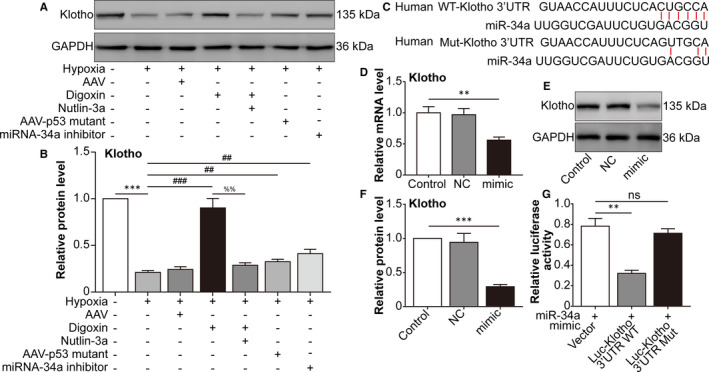
microrna‐34a inhibits the expression of Klotho. ARPE‐19 cells were divided into the following groups: normal, hypoxia, hypoxia + AAV vector, hypoxia + digoxin, hypoxia + digoxin + nutlin‐3a, hypoxia + AAV‐p53 mutant infection and hypoxia + miRNA‐34a inhibitor. A, Western blot assay of Klotho in ARPE‐19 cells was performed. B, The relative protein level of Klotho compared with the GAPDH level was analysed. *** P < .001, hypoxia group vs normal group. ### P < .001, ## P < .01, compared with the hypoxia group. %% P < .01, hypoxia + digoxin + nutlin‐3a group vs hypoxia + digoxin group. C, Schematic of the Klotho 3' UTR with the WT or mutated putative‐binding site of miRNA‐34a inserted into a luciferase (Luc) reporter. D, qRT‐PCR analysis of Klotho mRNA expression in HEK293T cells transfected with miRNA‐34a mimic for 24 h. ** P < .01, miRNA‐34a mimic group vs control group. E, Western blot of Klotho in HEK293T cells transfected with a miRNA‐34a mimic for 24 h was performed. F, Immunoblotted Klotho was quantified and normalized to GAPDH. *** P < .001, miRNA‐34a mimic group vs control group. G Relative luciferase activity was detected in HEK293T cells co‐transfected with plasmids containing firefly luciferase and wild‐type (WT) or mutant (Mut) Klotho 3′‐UTR and miRNA‐34a mimic for 24 h. The luciferase activity values were normalized to Renilla reniformis luciferase (TK‐RL) activity. ** P < .01, Luc‐Klotho 3′ UTR WT group vs vector group. Statistically non‐significant (NS), Luc‐Klotho 3′ UTR mutant group vs vector group
