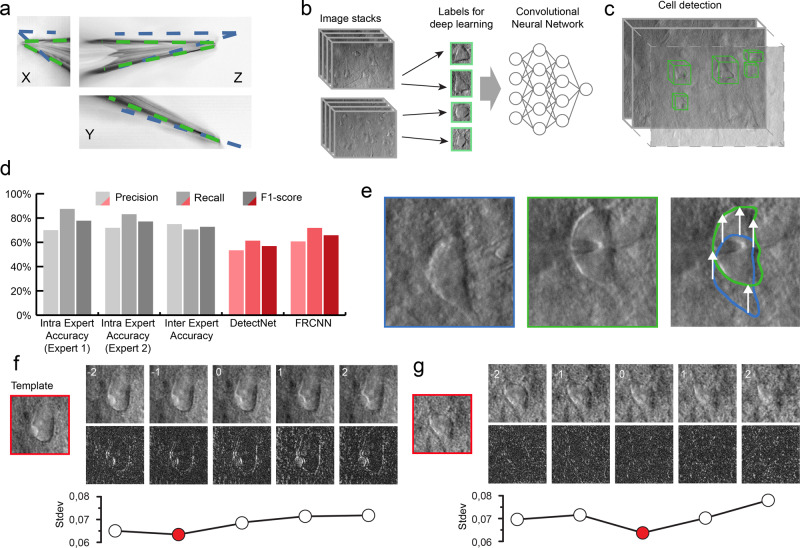
Fig. 3. The developed algorithms for the DIGAP system.
a Result of the Pipette Hunter detection model shown in three different projections of the image stack. Initial state (blue contour) and the result (green contour) of our pipette localization algorithm are shown. b Training dataset generation: 265 image stacks (60–100 images per stack with 1 μm frame distance along the Z-axis) captured from human and rodent neocortical slices with DIC videomicroscopy (left). 31,720 objects as healthy cells (green boxes) labeled on every slice of the image stack by four experts. c After the training session, the DIGAP system detects cells in unstained living neocortical tissues. d Accuracy of the automated cell detection pipeline. e Lateral tracking of the cell movement (n = 174). DIC images of the targeted (in blue box) and patched cell (in green box). The cell drifted from its initial location (arrows in the right panel) during the pipette maneuver. f, g Z-tracking of the cell movement (n = 174). The template image was captured at the optimal focal depth (in red boxes) before starting the tracking. During the pipette movement, image stacks were captured from the targeted cell (upper panels) such that the middle slice was taken of the most recent focus position. The bottom row shows the differences between the template and the image of the corresponding Z position. The lowest standard deviation value of the difference images (plots) shows the direction of the cell drift in the Z-axis. Source Data is available as a Source Data file.