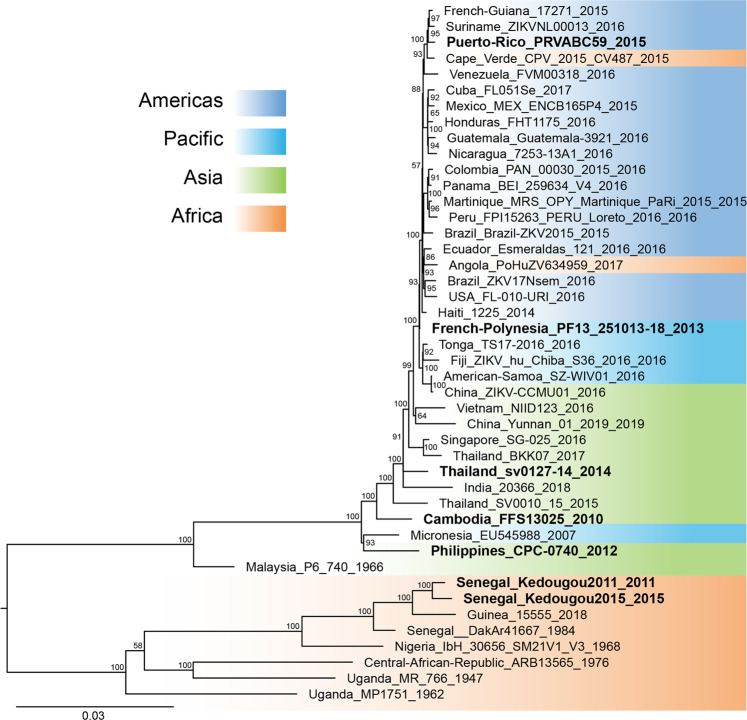
Fig. 1. Phylogenetic position of ZIKV strains used in this study.
The phylogenetic tree shows the seven ZIKV strains of the panel (in bold) among a backdrop of ZIKV strains spanning the current viral genetic diversity. The colored background represents the geographic origin of ZIKV strains. The consensus tree was generated from 1000 ultrafast bootstrap replicate maximum-likelihood trees, using a GTR + F + G4 nucleotide substitution model of the full ZIKV open reading frame. The tree is midpoint rooted and the root position is verified by the Spondweni virus outgroup on amino-acid and codon-based trees. Support values next to the nodes indicate ultrafast bootstrap proportions (%) and the scale bar represents the number of nucleotide substitutions/site.