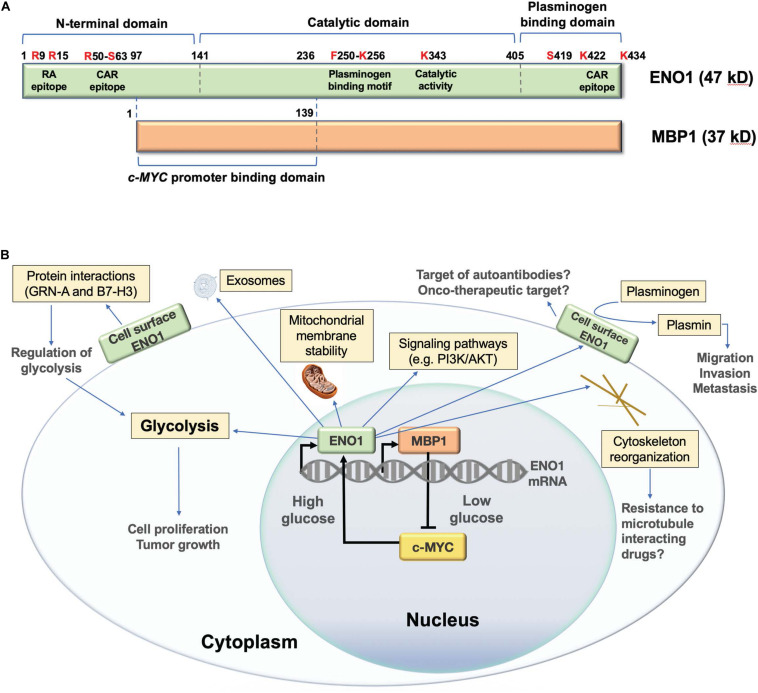
FIGURE 1.
Structure and functions of human ENO1. (A) Schematic representation of domain structure of ENO1 and its alternative translation variant c-MYC promoter binding protein 1 (MBP1). Several lysine residues (K256, K422, and K434) have been implicated in the plasminogen binding functions of ENO1, whereas K343 has been implicated in its catalytic activity, required for the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis. Citrullination of arginines 9 (R9) and 15 (R15) generates an immunodominant peptide (residues 5–22) that is targeted by autoantibodies in patients with rheumatoid arthritis. Methylation of arginine 50 (R50) has been implicated in ENO1 externalization. R50 is also part of an immunodominant epitope recognized by autoantibodies from patients with cancer associated retinopathy (CAR). Another CAR epitope is located within the plasminogen binding domain. Phosphorylated serine 419 (S419) within the plasminogen binding domain is recognized of ENO1 autoantibodies in pancreatic cancer patients. While MBP1 shares the catalytic and plasminogen binding domains of ENO1, it lacks these functions due to its exclusive nuclear localization. MPB1 residues 1–139 (ENO1 96–236) comprise its DNA binding domain, required for binding to the c-MYC gene promoter, which results in repression of promoter activity and downregulation of c-MYC protein expression. (B) Schematic representation of the cellular functions of ENO1 and MBP1. MBP1 localizes primarily in the cell nucleus where it represses the c-MYC gene promoter, whose activity is essential for ENO1 upregulation. ENO1 is primarily localized in the cytoplasm, where it functions in glycolysis, promoting mitochondrial stability and cytoskeleton reorganization, and regulating oncogenic signaling pathways. This protein is also localized on the cell surface, where it acts as a plasminogen receptor and interacting partner of various proteins to regulate glycolysis, as well as cancer cell migration, invasion, and metastasis. ENO1 can also be secreted from cells as a component of exosomal vesicles.