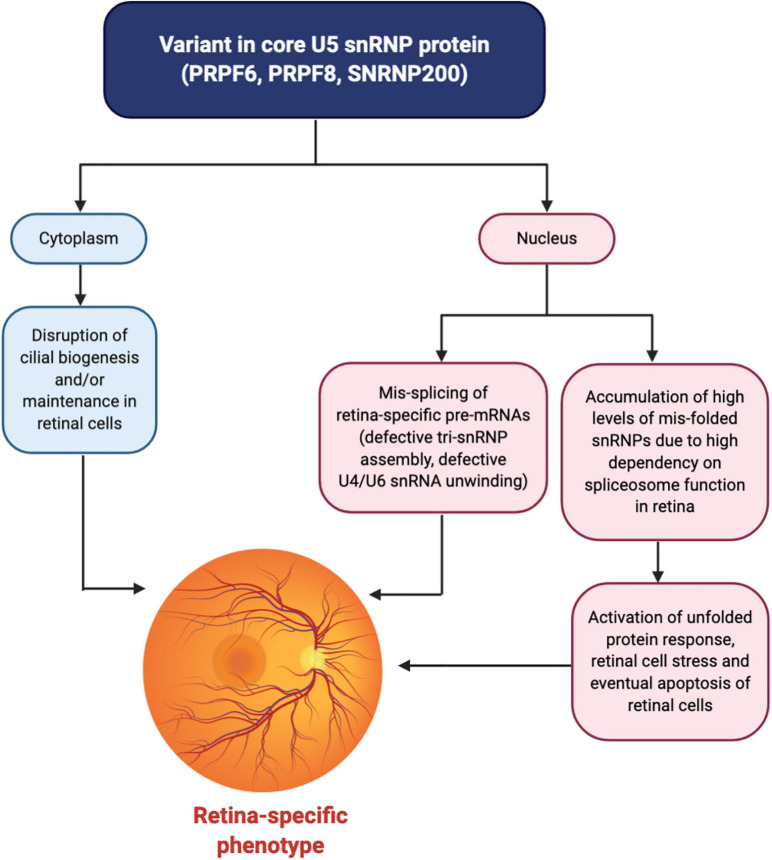
FIGURE 3.
Unified mechanism for a retinal-specific phenotype caused by variants in core U5 snRNP proteins. Variants affecting the PRPF6, PRPF8, or SNRNP200 proteins result in the mis-splicing of retina-specific pre-mRNAs in the nucleus through defective tri-snRNP assembly and/or defects in U4/U6 snRNA unwinding and spliceosome activation. Why the splicing of specific pre-mRNAs is affected is currently unknown although the pre-mRNAs may share common physical features. Furthermore, an accumulation of high levels of mis-folded snRNPs in the nucleus of retinal cells, largely stemming from the increased dependency on the spliceosome in the retina due to high levels of transcription and translation compared to other tissues, activates the unfolded protein response and generates cell stress. Over time, the accumulation of cell stress, along with photo-oxidative damage to the retinal cells, triggers apoptosis of retinal cells. In the cytoplasm, additional non-spliceosomal functions of these U5 snRNP proteins in cilia biogenesis and/or maintenance may be disrupted, affecting ciliated cells of the retina. These converging mechanisms together result in retinal degeneration and an eye-specific disease phenotype. Figure created with BioRender.com.