Fig. 4.
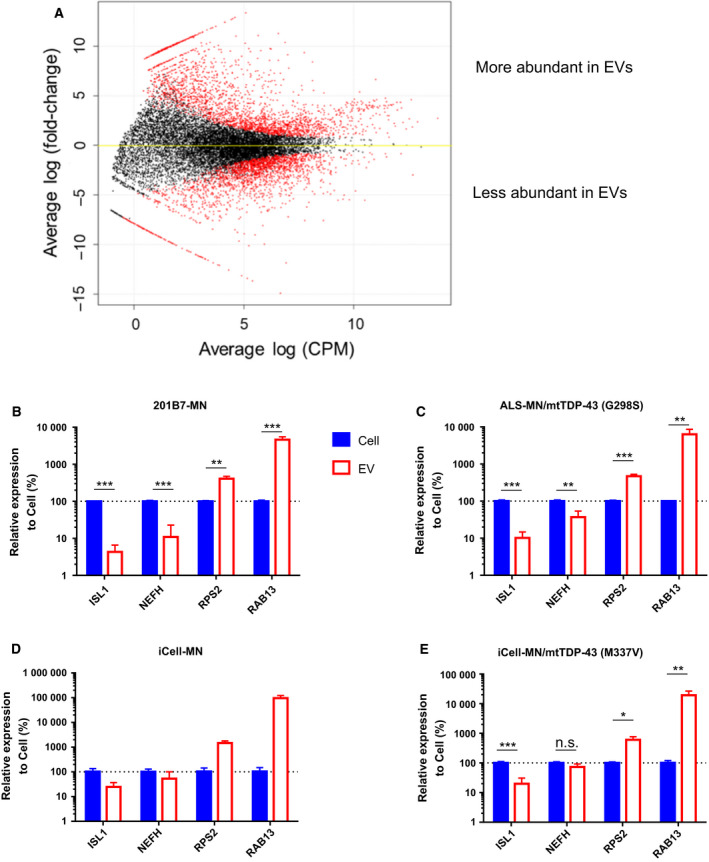
Differential analysis of gene expression between iPSC‐derived motor neurons and their EVs. (A) The result of differential analysis of gene expression by edgeR is visualized. The differentially expressed genes with adjusted P < 0.05 are colored in red. The representative differential abundance of ISL1, NEFH, RPS2 and RAB13 was confirmed by RT‐qPCR using several lines, (B) 201B7 iPSCs‐derived motor neurons, (C) ALS patient‐derived motor neurons harboring mutated TDP‐43 at G298S, (D) iCell motor neurons and (E) iCell motor neurons with genome‐edited TDP‐43 at M337V. The expression levels of these genes were normalized by ACTB. The data represent the average of n = 3 in (B), (C), and (E) and n = 2 in (D) with the error bar indicating SD. In statistical tests in (B), (C), and (E), t‐test without any assumption of variance was applied, and FDR for adjustment of multiple comparison was set to 0.05. The results were described as follows: ***P < 0.001, **P < 0.01, *P < 0.05; n.s., not significant. The statistical test was not performed in (D) because of insufficient number of replicates.
