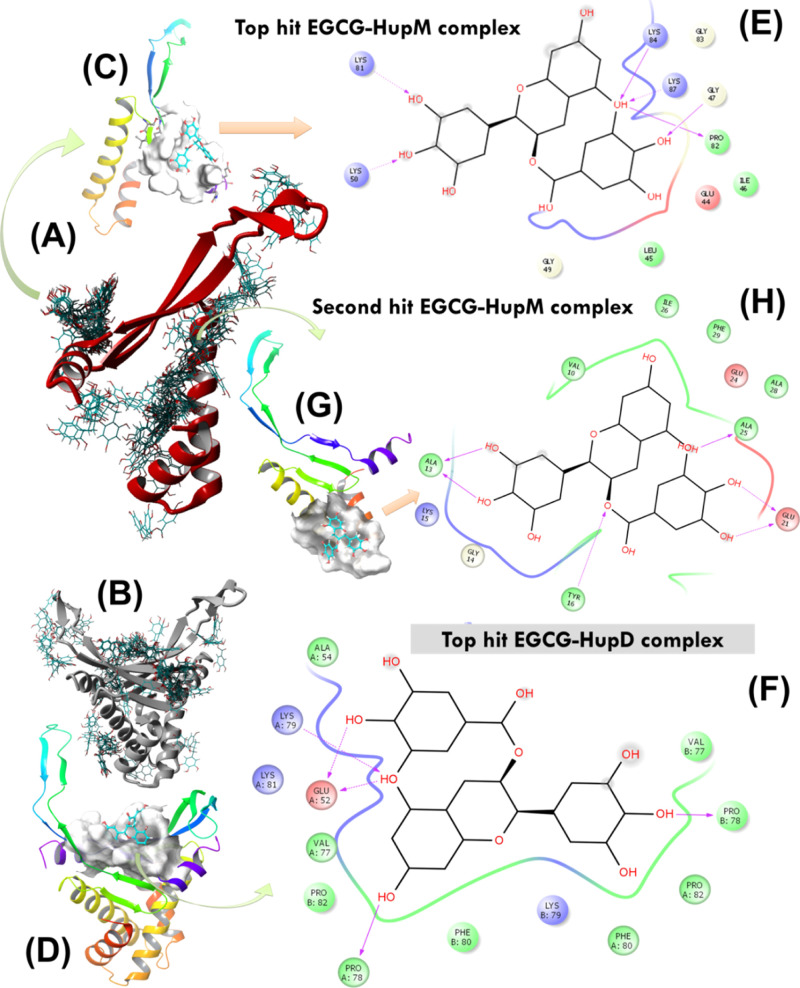
Figure 5.
(A,B) AUTODOCK molecular docking runs (N = 64) performed to search for potential EGCG binding sites over the protein surface of HupM and HupD. (C,D) Best docking poses of EGCG with HupM and HupD selected after cluster analysis based on highest binding energy of the complex. (E,F) 2D representation of molecular interaction for the ligand (EGCG) surrounded by contacting receptor residues. (G) Potential binding mode of EGCG with HupM selected after cluster analysis based on the second highest binding energy of the complex and the corresponding 2D representation of molecular interaction for the ligand (EGCG) surrounded by contacting receptor residues of HupM are shown in (H).