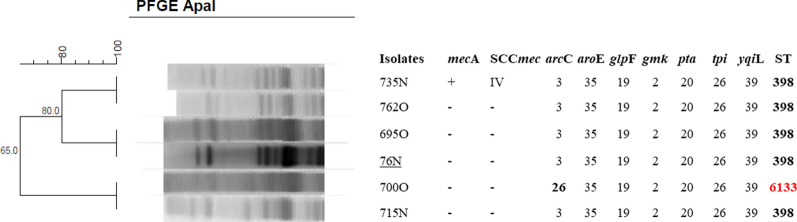
Fig. 3.
Dendrogram of the PFGE-ApaI profiles of MSSA isolates generated by Dice analysis/UPGMA (BioNumerics, Applied Maths) and sequence types obtained by MLST. Clustering of isolates digested with ApaI that were analyzed by MLST. All isolates except for 735 N were susceptible to methicillin (MSSA). N, nasal mucosa; O, oropharyngeal mucosa; arcC, carbamate kinase; aroE, shikimate dehydrogenase; glpF, glycerol kinase; gmk, guanylate kinase; pta, phosphate acetyltransferase; tpi, triosephosphate isomerase; yqiL, acetyl coenzyme A; ST, sequence type. Isolate 76 N was identified as ST398 in a previous study from our group. Isolate 700O was sent to the curator of the MLST database (https://pubmlst.org/) for identification of the new ST. This isolate was identified as ST 6133