Fig. 7 |. STF-SNPs perturb STF-DNA binding and abrogate signal responsiveness.
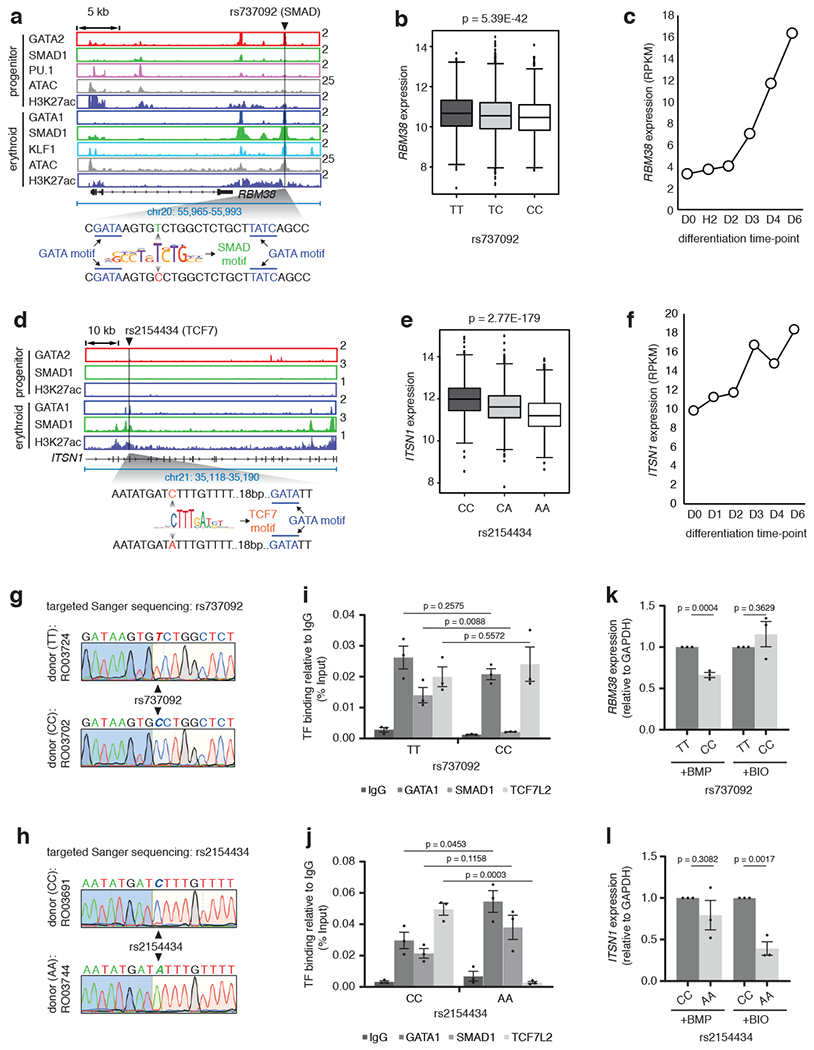
a, Gene tracks at RBM38 depicting the erythroid-specific TSC containing rs737092 at SMAD motif. b, RBM38 eQTL analysis for rs737092: boxplots represent median RBM38 expression as the thickest line, the first and third quartile as the box, and 1.5 times the interquartile range as whiskers. Two-sided test with linear model for EffectAlleleDosage used: effect estimate (β)=−0.05211; T-statistics=−13.6994, R2=0.034622; log10(P-value)=−41.2683 , log10(Benjamin-Hochberg’s FDR)=−38.6118. c, Expression RPKM values for the RBM38 gene at different stages of CD34+ erythroid differentiation. d, ITSN1 eQTL analysis for rs2154434: boxplots represent the median ITSN1 expression as the thickest line, the first and third quartile as the box, and 1.5 times the interquartile range as the whiskers. Two-sided test with linear model for EffectAlleleDosage used: effect estimate (β)=−0.0486; T-statistics=−29.7008, R2=0.144255; log10(P-value)=−178.558, log10(Benjamin-Hochberg’s FDR)=−175.322. f, Expression RPKM values for ITSN1 at stages of CD34+ erythroid differentiation. g and h, Sanger sequencing chromatograms of individual donors for SNPs rs737092 and rs2154434. Donor numbers indicated. i and j, Binding alteration of GATA1, SMAD1 and TCF7L2 for alternative alleles of rs737092 and rs2154434. Mean ± SEM shown. (n=3; 3 biologically independent experiments). Two-sided Students-t test used. k and l, qPCR analysis comparing the expression of RBM38 and ITSN1, relative to GAPDH, for alternative alleles of rs737092 and rs2154434, respectively, under BMP and BIO treatment. Mean ± SEM shown. (n=3; 3 biologically independent experiments). Two-sided Students-t test used.
