Abstract
Transforming growth factor (TGF)-β1 is a key cytokine affecting the pathogenesis and progression of cervical cancer. Tumor-derived exosomes contain microRNAs (miRNAs/miRs) that interact with cancer and stromal cells, thereby contributing to tissue remodeling in the tumor microenvironment (TME). The present study was designed to clarify how TGF-β1 affects tumor biological functions through exosomes released by cervical cancer cells. Deep RNA sequencing found that TGF-β1 stimulated cervical cancer cells to secrete more miR-663b-containing exosomes, which could be transferred into new target cells to promote metastasis. Further studies have shown that miR-663b directly targets the 3′-untranslated regions (3′-UTR) of mannoside acetylglucosaminyltransferase 3 (MGAT3) and is involved in the epithelial-mesenchymal transition (EMT) process. Remarkably, the overexpression of MGAT3 suppressed cervical cancer cell metastasis promoted by exosomal miR-663b, causing increased expression of epithelial differentiation marker E-cadherin and decreased expression of mesenchymal markers N-cadherin and β-catenin. Throughout our study, online bioinformation tools and dual luciferase reporter assay were applied to identify MGAT3 as a novel direct target of miR-663b. Exosome PKH67-labeling experiment verified that exosomal miR-663b could be endocytosed by cervical cancer cells and subsequently influence its migration and invasion functions which were measured by wound healing and Transwell assays. The expression of miR-663b and MGAT3 and the regulation of the EMT pathway caused by MGAT3 were detected by quantitative real-time transcription-polymerase chain reaction (qPCR) and western blot analysis. These results, thus, provide evidence that cancer cell-derived exosomal miR-663b is endocytosed by cervical cancer cells adjacent or distant after TGF-β1 exposure and inhibits the expression of MGAT3, thereby accelerating the EMT process and ultimately promoting local and distant metastasis.
Keywords: exosome, transforming growth factor-β1, miR-663b, mannoside acetylglucosaminyltransferase 3, epithelial-mesenchymal transition, cervical cancer
Introduction
Cervical cancer is the fourth most frequently diagnosed cancer and the fourth leading cause of cancer-related death in women with an estimated 570,000 cases and 311,000 deaths in 2018 worldwide (1,2). At present, the molecular mechanisms and precise targeted therapy of cervical cancer have attracted widespread research attention and provide new perspectives. Transforming growth factor (TGF)-β1 is a multifunctional regulatory peptide belonging to a newly discovered TGF-β superfamily that regulates cell growth, differentiation, proliferation and movement, widely involved in cell signal transduction in human cancers (3,4).
Exosomes are small disc-shaped membrane vesicles (30–150 nm) that contain complex RNA and proteins (5,6). Previous studies regard exosomes as specific secretory membrane vesicles involved in cell-to-cell communication (7,8). As the natural intercellular information carrier, exosomes are currently considered to have great potential in the field of drug carriers due to their small molecular structure, natural molecular transport characteristics and good biocompatibility (9,10). In addition, exosomes are increasingly regarded as exclusive media in the tumor microenvironment (TME) and molecular entities involved in its construction (11). Cancer-derived exosomes contain a variety of carcinogenic information that regulates the TME to promote the occurrence and progression of tumors.
Previous studies have confirmed that microRNAs (miRNAs/miRs) and other molecules are loaded to exosomes and secreted into the extracellular environment by many different cell types. MicroRNAs act as endogenous non-coding RNAs of approximately 20–24 nucleotides and key post-transcriptional regulators that affect gene expression. Through partial sequence complementation, they can bind to the 3′-untranslated regions (3′-UTR) of target mRNAs to cause mRNA degradation or translation inhibition (12). It was reported that miRNAs are widely involved in the regulation of cell differentiation, biological development and disease occurrence, as oncogenes or tumor-suppressor genes to affect the proliferation, migration and invasion of tumor cells (13–16). Epithelial-mesenchymal transition (EMT) plays a key role in cervical cancer and its extensive role in tumor progression has attracted more and more research attention (17). EMT refers to the biological process in which epithelial cells are transformed into mesenchymal phenotype cells, through which malignant tumor cells of epithelial origin gain the ability to migrate and invade (18–20).
Our present study demonstrated that TGF-β1 promotes the upregulation of exosomal miR-663b in cervical cancer cell lines by using deep RNA-seq. Further studies have shown that exosomes enriched in miR-663b could be endocytosed by recipient cells, causing the decreased expression of mannoside acetylglucosaminyltransferase 3 (MGAT3) and activation of the EMT signaling pathway, thereby promoting the metastasis of cervical cancer cells. Overall, our data provide new insight into the role of the TGF-β1/miR-663b/MGAT3 axis in cervical cancer.
Materials and methods
Cell lines and maintenance
293T cells and human cervical cell lines HeLa and CaSki were obtained from the Cancer Center Laboratory of Shandong University. 293T and HeLa cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) and CaSki cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS,; Biological Industries). All cell lines were cultured in a humidified incubator under standard culture conditions (5% CO2, 37°C).
Exosome isolation
Cells were seeded in 10-cm diameter plates and cultured in medium supplemented with 10% exosome-depleted FBS (System Biosciences). Culture medium was collected and exosomes were obtained using serial centrifugation. The medium was centrifuged at 300 × g for 10 min and 2,000 × g for 10 min to remove cells or other debris and 10,000 × g for 20 min to remove other larger vesicles. Then the medium was centrifuged at 110,000 × g for 70 min, washed with 5 ml phosphate-buffered solution (PBS) and centrifuged at 110,000 × g for another 70 min (all steps were performed at 4°C). Finally, the exosomes were obtained from the pellet and resuspended in PBS.
RNA sequencing (RNA-seq)
HeLa cells were seeded in a 10-cm diameter dish and cultured overnight, and exosome-free serum medium with or without 10 ng/ml TGF-β1 was used to continue the culture for another 24 h. The concentration of 10 ng/ml was much higher than the level of TGF-β1 secreted by tumor cells, thus, the impact produced by TGF-β1 secreted by tumor cells was negligible (21,22). The supernatant was collected to isolate exosomes and extract a total of 1 µg RNA as input material for RNA sample preparation. Total RNA was purified by electrophoretic separation on a 15% urea denaturing polyacrylamide gel electrophoresis (PAGE) gel and small RNA regions corresponding to the 18–30 nt bands in the marker lane (14–30 ssRNA ladder marker; Takara, Japan) were excised and recovered. Then the 18–30 nt small RNAs were ligated to a 5′-adaptor and a 3′-adaptor. The adapter-ligated small RNAs were subsequently transcribed into cDNA by SuperScript II Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) and then several rounds of PCR amplification with PCR Primer Cocktail and PCR Mix were performed to enrich the cDNA fragments. The PCR products were selected by agarose gel electrophoresis with target fragments 100–120 bp, and then purified using the QIA Quick Gel Extraction Kit (Qiagen). The library was evaluated for quality and quantitated using two methods: Checking the distribution of the fragment size using the Agilent 2100 bioanalyzer (Agilent Technologies, Inc.), and quantify the library using real-time quantitative PCR (qPCR) (TaqMan Probe). The final ligation PCR products were sequenced using the BGISEQ-500 platform. The RNA-sequence procedure and data analysis were completed by BGI (The Beijing Genomics Institute, BGI, ShenZhen, China).
Nanoparticle tracking analysis
Isolated exosomes diluted in PBS were gently mixed, and the mixture was inserted into the ZetaView PMX120 instrument (Particle Metrix GmbH) at 23°C through a 1-ml syringe. ZetaView 8.02.28 software (Particle Metrix GmbH) was utilized to analyze the number and the size distribution of the exosomes. Each sample was measured three times and the average value was calculated.
Transmission electron microscopy (TEM)
Isolated exosomes fixed with 2% paraformaldehyde for 5 min were dropped onto the Formvar copper carbonate grid with glow discharge for 1 min, and then it was negatively stained with 2% uranyl acetate for another 1 min. After sample drying, a HT7800 electron microscope (Hitachi) was used to photograph the grid with an acceleration voltage of 80 kV.
PKH67 staining for exosomes
PKH67 Green Fluorescent Cell Linker Kits (Sigma-Aldrich; Merck KGaA) were purchased to label the lipid bilayers of exosomes. PKH67 (4 µl) was added and exosomes were isolated into 1 ml diluent C separately and incubation was carried out for 5 min at room temperature; exosomes without PKH67 staining were used for negative control. Bovine serum albumin (1 ml/5%) (Solarbio) was added to halt the staining. Then the PKH67-labeled exosome mixture was centrifuged at 110,000 × g for 2 h at 4°C. The mixture was washed with PBS and centrifuged at 110,000 × g for another 2 h to obtain pure PKH67-labeled exosomes. After resuspending the mixture in complete medium and incubating with HeLa cells at 37°C for 0, 4, 8 and 12 h, a laser confocal microscope (LSM880; Zeiss) was used to visualize the incorporation of exosomes into HeLa cells.
Western blot analysis
Protein extracted from the cells and exosomes was lysed with RIPA buffer and quantified by BCA Protein Assay Kit (Beyotime Institute of Biotechnology). Total proteins were separated on a 10% polyacrylamide gel and transferred to a polyvinylidene fluoride membrane and blocked with 5% defatted milk for 1.5 h at room temperature. The membranes were probed with the following primary antibodies at 4°C: Anti-CD63 (dilution 1:1,000, Abcam, ab59479), anti-TSG101 (dilution 1:1,000; Abcam, ab125011), anti-HSP70 (dilution 1:2,000; ProteinTech Group, Inc., 66183-1-Ig), anti-GAPDH [dilution 1:1,000; Cell Signaling Technology, Inc. (CST), #5174], anti-MGAT3 (dilution 1:1,000; ABclone, A8134), anti-E-cadherin (dilution 1:1,000; CST, #3195), anti-N-cadherin (dilution 1:1,000; CST, #13116), and anti-β-catenin (dilution 1:1,000; CST, #8480). Subsequently, the membranes were washed with triethanolamine buffered saline solution (TBS) and interacted with HRP-conjugated antibody (dilution 1:1,000; CST) for 2 h at room temperature. The membranes were detected by ECL substrate kit (Thermo Fisher Scientific Inc.) and quantified by ImageJ software (v1.8.0; National Institutes of Health, USA).
RNA isolation and quantitative real-time transcription-polymerase chain reaction (qPCR)
Total RNA extracted from cells or exosomes was isolated with TRIzol reagent (Thermo Fisher Scientific Inc.) according to the manufacturer's instructions and the concentration was detected by a spectrophotometer (Thermo Fisher Scientific Inc.). Then the RNA was transcribed to cDNA by using Prime Script RT Master Mix for RT-PCR (Takara). qPCR was conducted using SYBR-Green (Takara) and performed on a StepOne™ PCR amplifier (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 were used as internal controls. The primer sequences for miRNA and mRNA were purchased from GenePharma (Shanghai, China) and are as follows: miR-663b, forward (F), 5′-GGTGGCCCGGCCGTGC-3′ and reverse (R), 5′-TATCCTTGTTGACGACTCCTTGAC-3′; U6, F, 5′-CAGCACATATACTAAAATTGGAACG-3′ and R, 5′-ACGAATTTGCGTGTCATCC-3′; MG AT3, F, 5′-TCAACCACGAGTTCGACCTG-3′ and R, 5′-CACCTTGTGGCGGATGTACT-3′; GAPDH, F, 5′-GCACCGTCAAGGCTGAGAAC-3′ and R, 5′-TGGTGAAGACGCCAGTGGA-3′.
Wound healing assay
Cells were seeded in 6-well plates until the growth density reached 90%. Then a sterile 100-µl pipette tip was used to quickly stroke the cells to form a straight wound. After washing twice with PBS, 2 ml medium without serum was added to each well. The wound healing was observed and photographed by JEM-1200 EX II electron microscope (JEOL) at 0 and 24 h, and the wound closure was evaluated by ImageJ software (v.8.0; National Institutes of Health).
Transwell assays
Cells (HeLa: 6×104 cells/200 µl for invasion and 4×104 cells/200 µl for migration, CaSki: 8×104 cells/200 µl for invasion and 6×104 cells/200 µl for migration) in 200 µl serum-free medium were seeded in upper Transwell chambers (8-µm pore size; Corning Costar) with or without Matrigel (60 µl, 1:9 dilution in serum-free medium, BD Biosciences). In addition, 600 µl media containing 10% FBS were added to the lower compartment. After 24 h of incubation in an incubator, the chambers were washed twice by PBS, and methanol was used to fix cells for 15 min. The cells that did not pass through the upper layer were wiped off with a cotton swab and then stained with crystal violet (product no. AC0121; Beyotime) for 30 min. A total of 3–5 random fields of view were selected to photograph under the JEM-1200 EX II electron microscope (JEOL).
miRNA target prediction and dual luciferase reporter assay
PicTar (https://pictar.mdc-berlin.de/), TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do) were used to predict miR-663b targets. The expression of the miR-663b-target gene was verified in 293T cells by dual luciferase reporter gene assay. First, we insert the 3′-UTR of wild-type (WT) and mutant (MUT) MGAT3 into the psiCHECKTM-2 plasmid (Promega Corp.). Then, psiCHECKTM-2-MGAT3-3′-UTR-WT or psiCHECKTM-2-MGAT3-3′-UTR-MUT were co-transfected into 293T cells with miR-663b mimics or its NC. After 48 h of transfection in a 96-well plate, a Dual Fluorescence Luciferase reporter gene detection system (Promega Corp.) was used to detect the fluorescence.
Cell transfection
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific Inc.) and Opti-MEM (Gibco; Thermo Fisher Scientific Inc.) were used for cell transfection. miR-663b mimics and inhibitor were used for upregulation and downregulation of miR-663b. miR-663b mimics NC and inhibitor NC were non-targeting and used as the respective controls; the control (CON) group was without any special treatment. The overexpression of MGAT3 was produced by subcloning PCR-amplified full-length human MGAT3 cDNA into the pCDNA3.1 plasmid (GenePharma). The empty vector was used as a blank control (pCDNA3.1 NC). After transfection in Opti-MEM for 6 h, completed culture medium was replaced and the cells were cultured for the following experiments. Total RNA and total protein were isolated from the cells 24 or 48 h after transfection for qPCR analysis and western blot analysis. In order to obtain stably silenced cell lines for subsequent experiments, we used the lentiviral vector miR663b-sponge-pLVX-AcGFP-N1-Puro to transfect HeLa and CaSki cells and cultured the cells with puromycin dihydrochloride (2 µg/ml; Amresco) for 7–10 days. The intensity and ratio of green fluorescence produced by cells under the fluorescence microscope were used to prove the efficiency of lentiviral transfection.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 8 software (GraphPad Software, Inc.). The data are expressed as the median values ± standard deviation (SD). Comparisons between the groups were analyzed by Student's t-test (unpaired, two-tailed) or one-way analysis of variance (ANOVA) followed by Tukey's post hoc test. The results are presented as the average of three experiments. Statistical significance was set at P<0.05.
Results
Elevated exosomal miRNAs upon TGF-β1 exposure of cervical cancer cell lines are identified by deep RNA-sequencing (RNA-seq)
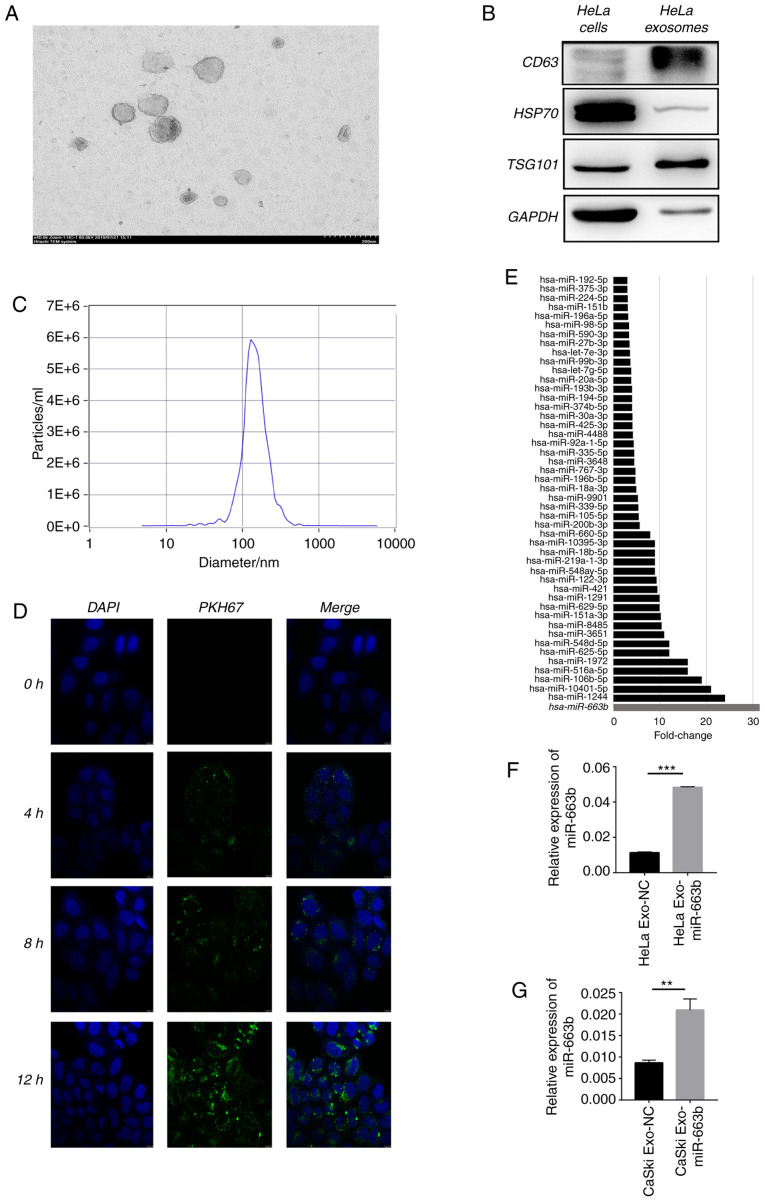
First, deep RNA-seq was performed to explore the expression level of miRNAs in HeLa cell exosomes after treatment with or without 10 ng/ml TGF-β1 for 24 h. The typical goblet morphology and the size range of 30–150 nm were detected by TEM (Fig. 1A). Next, positive signs of exosomes including CD63, tumor susceptibility gene 101 (TSG101), heat shock protein 70 (HSP70) and GAPDH were confirmed by western blot analysis (Fig. 1B). NTA proved that the concentration and size of the isolated exosomes were consistent with previous reports (Fig. 1C) (23). The green fluorescent signal in the HeLa cytoplasm indicated that the purified exosomes labeled with the fluorescent membrane tracer PKH67 (green) entered the HeLa cells after incubation (Fig. 1D). Deep RNA-seq results showed that 48 miRNAs were enriched in exosomes of HeLa cells after treatment with TGF-β1 compared with the non-treated cells (negative control) (Fig. 1E), proving that TGF-β1 affects the HeLa cell exosome miRNA profile. We selected top 10 miRNAs for PCR verification in HeLa and CaSki cells. After treatment of TGF-β1 for 24 h, miR-663b was selectively enriched 4 and 3 times in the exosomes of HeLa and CaSki cells compared with the negative control group (Fig. 1F and G). These results verified that miR-663b could be selectively enriched in the exosomes of cervical cancer cells treated with TGF-β1, thus we selected miR-663b for the subsequent study.
Figure 1.
Elevated exosomal miRNAs upon TGF-β1 exposure of cervical cancer cell lines are identified by deep RNA-sequencing (RNA-seq). (A) TEM images of exosomes isolated from HeLa cells. (B) HeLa cell-secreted exosome-positive markers CD63, TSG101, HSP70 and GAPDH were detected by western blot analysis. (C) Nanoparticle size analysis of HeLa cell-secreted exosomes. (D) HeLa cells pretreated with PKH67-labeled exosomes for 0, 4, 8 and 12 h were stained by DAPI (blue) for confocal microscopy analysis (magnification, ×400). (E) miR-663b expression level in HeLa-secreted exosomes with or without TGF-β1 treatment were identified by deep RNA-seq. (F and G) miR-663b expression level in HeLa and CaSki-secreted exosomes with or without TGF-β1 treatment were analyzed by qPCR. **P<0.01, ***P<0.001. Exo-miR-663b, exosomes from the TGF-β1 treatment group; Exo-NC, exosomes from the negative control group. TGF-β1, transforming growth factor-β1; TEM, transmission electron microscopy; TSG101, tumor susceptibility gene 101; HSP70, heat shock protein 70.
Exosomal miR-663b promotes the metastasis of cervical cancer cells
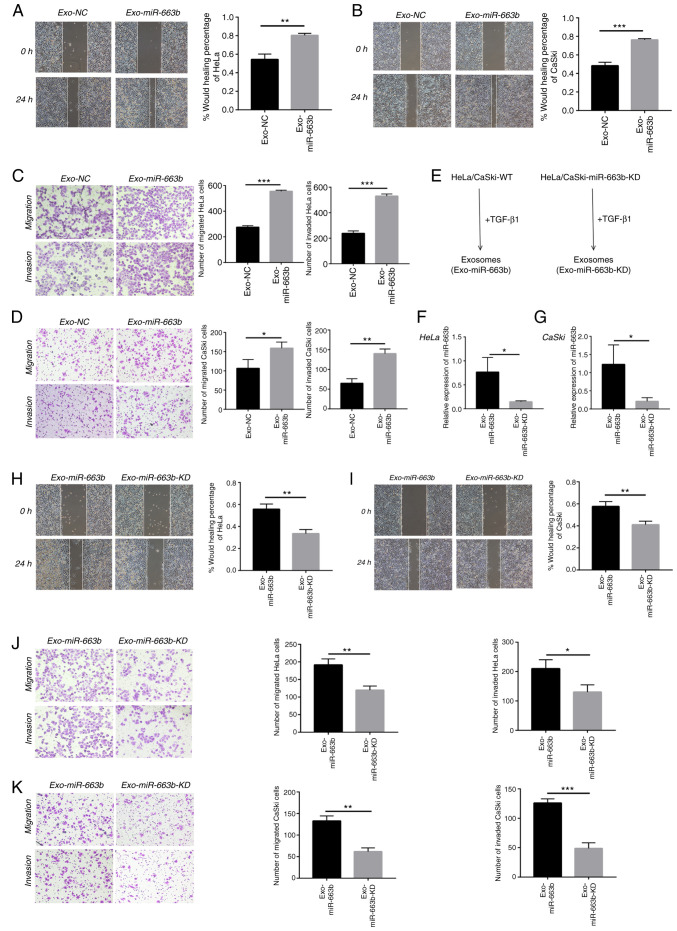
Previous research has confirmed that miR-663b plays tumor-promoting roles in endometrial cancer, nasopharyngeal carcinoma and osteosarcoma (24–26). We collected exosomes from the TGF-β1 treatment group (Exo-miR-663b) and the negative control group (Exo-NC) in HeLa and CaSki cells to verify whether exosomal miR-663b affects migration, invasion or proliferation of cervical cancer cells. In our study, HeLa and CaSki cells were treated with exosomes from their own cells. We found exosomes from the TGF-β1 treatment group significantly enhanced HeLa and CaSki cell motility by wound healing assay (Fig. 2A and B). In addition, exosomes upon TGF-β1 exposure enhanced HeLa and CaSki cell migration and invasion as measured by Transwell assays (Fig. 2C and D).
Figure 2.
Exosomal miR-663b promotes the metastasis of cervical cancer cells. (A) HeLa cell migration with normal or TGF-β1-treated exosomes was assessed by wound healing assay. (B) CaSki cell migration with normal or TGF-β1 treated exosomes was assessed by wound healing assay. (C) Migration and invasion abilities of HeLa cells with normal or TGF-β1-treated exosomes were detected by Transwell assays. (D) Migration and invasion abilities of CaSki cells with normal or TGF-β1-treated exosomes were detected by Transwell assays. (E) Schematic diagram of the exosomes-tumor cell experiments. (F and G) miR-663b expression levels in HeLa and CaSki cell-secreted exosomes from the Exo-miR-663b and Exo-miR-663b-KD group were analyzed by qPCR. (H) Wound healing assays were used to detect HeLa cell migration after treatment with exosome from miR-663b-KD or WT cells upon TGF-β1 exposure. (I) Wound healing assays were used to detect CaSki cell migration after treament with exosome that from miR-663b-KD or WT cells upon TGF-β1 exposure. (J) Transwell assay was performed to assess the effect of miR-663b-KD or WT exosomes following exposure of TGF-β1 on migration and invasion of HeLa cells. (K) Transwell assay was performed to measure the effect of miR-663b-KD or WT exosomes following exposure of TGF-β1 on migration and invasion of CaSki cells. Magnification, ×100, *P<0.05, **P<0.01, ***P<0.001. Exo-miR-663b, exosomes from the TGF-β1 treatment group; Exo-NC, exosomes from the negative control group; Exo-miR-663b-KD, exosomes from the miR-663b-knockdown group. TGF-β1, transforming growth factor-β1; MGAT3, mannoside acetylglucosaminyltransferase 3; WT, wild-type.
To further explore that exosomal miR-663b is indeed involved in cervical cancer cell metastasis, we used a lentiviral vector to knock down miR-663b (Exo-miR-663b-KD). The intensity and ratio of green fluorescence were used to prove the efficiency of lentiviral transfection and the images are provided in Fig. S1. After treatment with TGF-β1, we collected exosomes from the medium of the Exo-miR-663b-KD group and wild-type group (Exo-miR-663b) (Fig. 2E). qPCR verified that the amount of miR-663b in exosomes from the Exo-miR-663b group was much higher than that in the Exo-miR-663b-KD group both in HeLa and CaSki cell lines (Fig. 2F and G). Then we observed that compared with the Exo-miR-663b treatment group, the ability of migration and invasion of HeLa and CaSki cells was attenuated when the Exo-miR-663b-KD was incubated (Fig. 2H-K). Together, our results indicated that exosomes enriched in miR-663b after TGF-β1 treatment could be transferred into new target cells to promote the metastasis of cervical cancer.
Exosomal miR-663b directly targets MGAT3
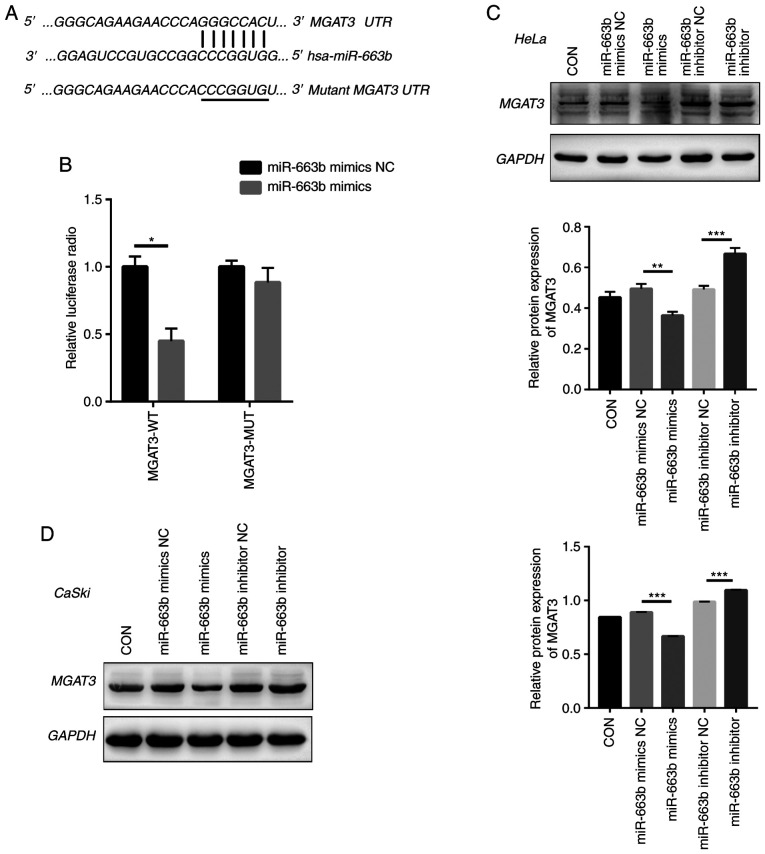
Next, we used bioinformatic tools (TargetScan, PicTar, and miRanda) to explore the potential miR-663b target genes. We identified a 7-bp binding site between MGAT3 3′-UTR and miR-663b (Fig. 3A). MGAT3, as a key glycosyltransferase in the N-glycan biosynthetic pathway (27), is considered to be a metastasis suppressor gene (28), which regulates the glycosylation step and the key tumor-suppressor gene E-cadherin to inhibit tumor invasion and metastasis (29–31). To confirm that miR-663b could regulate the putative target of MGAT3, the predicted miR-663b binding site (wild-type) or mutant sequence (mutant type) in the 3′-UTR of MGAT3 was cloned into a luciferase reporter plasmid and we detected the response to miR-663b in 293T cells. Our results showed that the co-transfection of miR-663b mimics significantly reduced the luciferase activity of the wild-type 3′-UTR of MGAT3, while the luciferase activity of the mutant 3′-UTR MGAT3 had no change (Fig. 3B).
Figure 3.
Exosomal miR-663b directly targets MGAT3. (A) The putative binding site between MGAT3 and miR-663b. (B) Luciferase activity was detected in 293T cells co-transfected with miR-663b mimics or miRNA NC and WT-MGAT3 or MUT-MGAT3. (C) The expression of MGAT3 in HeLa cells transfected with miR-663b mimics, miR-663b inhibitor and NC groups were detected by western blot analysis. (D) The expression of MGAT3 in CaSki cells transfected with miR-663b mimics, miR-663b mimics inhibitor and NC groups were detected by western blot analysis. *P<0.05, **P<0.01 and ***P<0.001. CON, control; NC, negative control; MGAT3, mannoside acetylglucosaminyltransferase 3; WT, wild-type; MUT, mutant.
Moreover, we directly transfected HeLa and CaSki cells with miR-663b mimics, miR-663b inhibitor, miR-663b mimics NC and miR-663b inhibitor NC, respectively. Consistent with the results of dual luciferase reporter gene assay, compared with the NC group, miR-663b mimics resulted in decreased MGAT3 protein expression in HeLa and CaSki cells (Fig. 3C and D), while treatment with miR-663b inhibitor resulted in increased MGAT3 protein expression. Above all, our results demonstrated that exosomal miR-663b could inhibit MGAT3 expression.
MGAT3 is involved in cervical cancer metastasis promoted by exosomal miR-663b
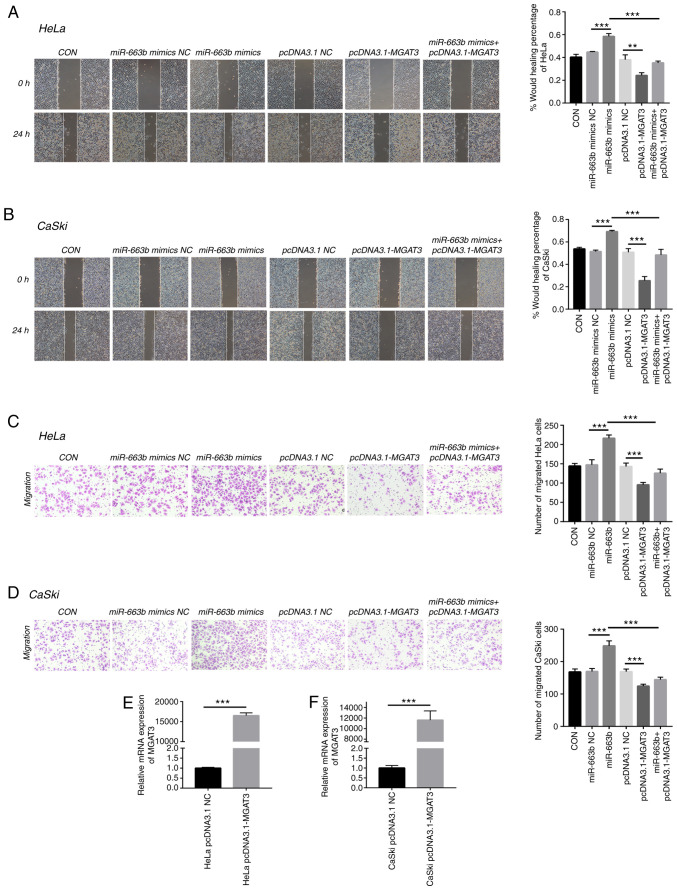
To further confirm the role of MGAT3 in cervical cancer cells, we explored the function of exosomal miR-663b on cell migration under the vector-mediated MGAT3 gene overexpression conditions. Wound healing and Transwell assays were performed to show that miR-663b could enhance the migration and invasion ability of HeLa and CaSki cells, while co-transfection with pcDNA3.1-MGAT3 partially alleviated this effect caused by miR-663b (Fig. 4A-D). qPCR was used to verify the overexpression efficiency of MGAT3 plasmid transfected into HeLa and CaSki cells (Fig. 4E and F). These results indicated that exosomal miR-663b promoted the metastasis ability of cervical cancer cells by inhibiting the expression of MGAT3.
Figure 4.
MGAT3 is involved in cervical cancer metastasis promoted by exosomal miR-663b. (A) Wound healing assays were performed to detect the migratory ability of HeLa cells transfected with miR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 (magnification, ×100). (B) Wound healing assays were performed to detect the migratory ability of CaSki cells transfected with miR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 (magnification, ×100). (C) Transwell assay was performed to detect the effect of miR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 on the migration of HeLa cells (magnification, ×100). (D) Transwell assay was performed to detect the effect of miR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 on the migration of CaSki (magnification, ×100). (E) The mRNA level of MGAT3 in HeLa cells after transfection with pCDNA3.1-MGAT3 and pCDNA3.1 NC plasmid were analyzed by qPCR. (F) The mRNA level of MGAT3 in CaSki cells after transfection with pCDNA3.1-MGAT3 and pCDNA3.1 NC plasmid were analyzed by qPCR. **P<0.01 and ***P<0.001. CON, control, MGAT3, mannoside acetylglucosaminyltransferase 3; NC, negative control.
MGAT3 inhibits metastasis ability of cervical cancer by affecting the EMT pathway
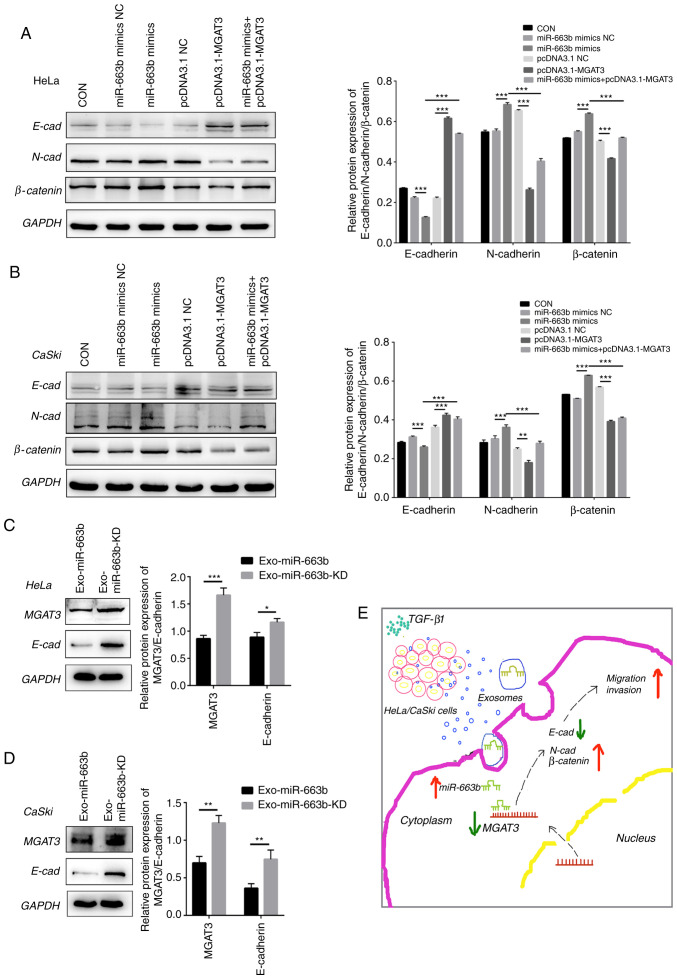
As the key role in cancer recurrence and exosome-mediated tumor progression, EMT is a biological process in which epithelial cells transform into cells with a mesenchymal phenotype and cause increased invasiveness and subsequent metastasis (18,20). To understand the potential molecular mechanism of MGAT3 in inhibiting cervical cancer invasion and metastasis, we detected the level of epithelial differentiation marker E-cadherin and mesenchymal marker N-cadherin and β-catenin in cervical cancer cells. Western blot analysis verified that after transfected with pcDNA3.1-MGAT3, the expression of E-cadherin was significantly increased in HeLa cells, while the expression of N-cadherin and β-catenin were significantly decreased. In contrast, after transfection with the miR-663b mimics, the expression of N-cadherin and β-catenin were significantly increased, while the expression of E-cadherin was significantly decreased. Moreover, all of these effects caused by miR-663b could be offset by the overexpression of MGAT3 in vitro (Fig. 5A). We also observed the same results in CaSki cells (Fig. 5B).
Figure 5.
MGAT3 inhibits the metastatic ability of cervical cancer cells by affecting the EMT pathway. (A) miR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 were transfected into HeLa cells, respectively. Western blot analysis was performed to analyze E-cadherin (E-cad), N-cadherin (N-cad) and β-catenin expression. (B) MiR-663b mimics NC, miR-663b mimics, pcDNA3.1 NC, pcDNA3.1-MGAT3 and miR-663b mimics + pcDNA3.1-MGAT3 were transfected into CaSki cells, respectively. Western blot analysis was performed to analyze E-cad, N-cad and β-catenin expression. (C) Western blot analysis was performed to analyze MGAT3, E-cad, N-cad and β-catenin expression in HeLa cells in the Exo-miR-663b group and Exo-miR-663b-KD group. (D) Western blot analysis was performed to analyze MGAT3, E-cad, N-cad and β-catenin expression in CaSki cells in the Exo-miR-663b group and Exo-miR-663b-KD group. (E) Schematic representation of the TGF-β1/miR-663b/MGAT3 axis in cervical cancer. *P<0.05, **P<0.01 and ***P<0.001. Exo-miR-663b, exosomes from the TGF-β1 treatment group; Exo-miR-663b-KD, exosomes from the miR-663b-knockdown group. EMT, epithelial-mesenchymal transition; CON, control, MGAT3, mannoside acetylglucosaminyltransferase 3; TGF-β1, transforming growth factor-β1; NC, negative control.
In order to further confirm that exosomal miR-663b promotes metastasis by inhibiting the expression of MGAT3 and affecting the EMT pathway, we evaluated the expression of MGAT3 and E-cadherin after the treatment of the cell lines with Exo-miR-663b and Exo-miR-663b-KD. Compared with the Exo-miR-663b-KD group, the expression of MGAT3 in the Exo-miR-663b group was significantly decreased, accompanied by the decreased expression of E-cadherin (Fig. 5C and D). The underlying mechanism of the TGF-β1/miR-663b/MGAT3 axis in cervical cancer is shown in Fig. 5E.
Discussion
Accumulated research has shown that exosomes can mediate cell-to-cell communication (32,33), package and transport microRNAs to new cells and participate in the regulation of gene expression. MicroRNAs in exosomes are being suggested as novel biomarker for cervical cancer prediction and diagnosis (34). In the present study, RNA-seq analyzed miRNA profiles of HeLa and CaSki exosomes and verified that miR-663b was preferentially enriched in exosomes following treatment with TGF-β1. qPCR was performed to confirm the expression of miRNAs in HeLa and CaSki exosomes upon TGF-β1 exposure. TGF-β1 exerts growth inhibition and displays an anti-inflammatory function in homeostasis and early stages of cancer (35,36), while abnormal TGF-β activation in the advanced stage of tumors promotes aggressive growth characteristics and metastatic spread (37). Previous studies have reported that the TGF-β1 pathway regulates miRNA expression, but its role in cervical cancer is controversial (38). The concentration of 10 ng/ml that was used in this study was much higher than the level of TGF-β1 secreted by tumor cells and can make the effect of TGF-β1 secreted by itself negligible (21,22). miR-663b was reported to be elevated in endometrial cancer, nasopharyngeal carcinoma, and osteosarcoma, and its expression is associated with cell invasion, apoptosis, and chemotherapy resistance (24–26). miR-663b may be epigenetically repressed by pterostilbene in human endometrial cancer cells and repressed by long non-coding RNA HOTAIR and exerts its tumor-suppressive function via targeting insulin-like growth factor 2 in pancreatic cancer (39,40). However, the expression and regulatory mechanism of secreted miR-663b in cervical cancer remain unclear, which may be the future direction of exosomal research. To explore the role of exosomal miR-663b in the progression of cervical cancer, exosomes collected from TGF-β1-treated and non-treated cells were incubated with HeLa and CaSki cells, and wound healing and Transwell assays were preformed to measure cell migration and invasion ability. Our results revealed that miR-663b could be enriched in exosomes upon TGF-β1 exposure and transported into new target cells to promote cervical cancer cell metastasis. However, flow cytometry, CCK-8 and EdU assays showed that there was no significant difference in apoptosis and proliferation ability among these two groups (data not shown). Our data seem to provide a hypothesis that exosomal miRNAs can amplify metastatic elements into cells that are adjacent or distant to effectively develop tumors.
miRNAs cleave target mRNAs or regulate target gene protein expression through perfect or nearly perfect complementary binding to the 3′-UTR or open reading frame (ORF) region of the target genes (41). In the present study, dual luciferase activity assay showed that exosomal miR-663b directly targets the 3′-UTR of mannoside acetylglucosaminyltransferase 3 (MGAT3), and western blot analysis confirmed that the protein level of MGAT3 was decreased in recipient cells. Previous studies have reported that glycosyltransferase MGAT3 is closely related to cell proliferation, migration and metastasis in a variety of cancers (42–48). MGAT3 can catalyze the transfer of β-N-acetylglucosamine (GlcNAc) in the β-1,4 bond to mannose on the N-glycan, thereby inhibiting the β1-6GlcNAc formation catalyzed by another glycosyltransferase MGAT5. β1-6GlcNAc is frequently detected to be overexpressed in a variety of metastatic tumors (49,50). Metastatic ovarian cancer and lung cancer have been reported to exhibit significant reductions in mRNA and protein levels of MGAT3 (51). In our study, it was found that the overexpression of MGAT3 effectively inhibited HeLa and CaSki cell ability for migration and invasion. Taken together, our data indicate that miR-663b promotes the metastasis of cervical cancer cells by inhibiting MGAT3 activity in the N-glycan pathway of cervical cancer cells.
In previous studies, MGAT3 was shown to affect the activation of signal transduction pathways by regulating extracellular signal-regulated kinase (ERK)1/2 or protein kinase B (AKT) signals (52), and was involved in EMT progression and its reverse processes (42,53). Therefore, we examined the possible consequences of abnormal MGAT3 expression on related signaling pathways in cervical cancer. In the miR-663b mimics group, it was observed that E-cadherin expression was decreased and N-cadherin and β-catenin were increased. In the pcDNA3.1-MGAT3 group, MGAT3 overexpression had a significant blocking effect on the downregulation of E-cadherin and the upregulation of N-cadherin and β-catenin. These findings together demonstrated that the overexpression of MGAT3 inhibited the EMT process in cervical cancer cell lines.
Multiple factors such as pterostilbene or long non-coding RNAs that could affect the expression of miR-663b were not investigated in our present study. This was the limitation of our current research, but it also provides us with possible directions and effective goals for future research. We will further explore more factors affecting miR-663b expression and clarify whether exosomal miR-663b could be used to predict the recurrence and metastasis in cervical cancer patients, and evaluate the effectiveness of miR-663b as a new type of targeted therapy.
In our current study, we provide evidence that TGF-β1 can selectively promote the expression of miR-663b in cervical cancer exosomes. After being transported to new target cells via exosomes, miR-663b significantly inhibits the expression of MGAT3 and enhances the metastatic ability of cervical cancer cells. We will conduct more precise research to support our results.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This study was carried out at Qilu Hospital of Shandong University and was supported by the Shandong Provincial Key Research Project (2017CXGC1210, 2019GSF108126), the National Natural Science Foundation of China (NSFC, 81572559, 81902644), the Natural Science Doctoral Program Foundation of Shandong Province (ZR2019BC059) and the Health Commission of Weifang (wfwsjs-2018-053).
Funding
This study was carried out at Qilu Hospital of Shandong University and was supported by the Shandong Provincial Key Research Project (2017CXGC1210, 2019GSF108126), the National Natural Science Foundation of China (NSFC, 81572559, 81902644), the Natural Science Doctoral Program Foundation of Shandong Province (ZR2019BC059) and the Health Commission of Weifang (wfwsjs-2018-053).
Availability of data and materials
All data used in this study can be obtained from the corresponding author upon reasonable request. The RNA sequencing data has been uploaded to the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with data number GSE163507.
Authors' contributions
YZ was mainly responsible for project design and revision of important knowledge content. XY was responsible for the implementation of the experiment, data analysis, and wrote the initial version of the manuscript. YW conducted the statistical analysis, reviewed and edited the final manuscript. JM performed the experiments and analyzed data. SH conducted the statistical analysis, reviewed and edited the final manuscript. LL analyzed the data and revised the content. YS maintained the cells, analyzed the data and reviewed the manuscript. JZ, SS, XL, WS and YD analyzed and interpreted the data. All authors participated in this research and agreed to be responsible for all aspects of the research; they read and approved the final manuscript to ensure the accuracy and integrity of the work.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2018;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;23:77–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun D, Zhuang X, Zhang S, Deng ZB, Grizzle W, Miller D, Zhang HG. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Deliv Rev. 2013;65:342–347. doi: 10.1016/j.addr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 7.OBrien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21:585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurywchak P, Kalluri R. An evolving function of DNA-containing exosomes in chemotherapy-induced immune response. Cell Res. 2017;27:722–723. doi: 10.1038/cr.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Veerman RE, Güçlüler Akpinar G, Eldh M, Gabrielsson S. Immune cell-derived extracellular vesicles-functions and therapeutic applications. Trends Mol Med. 2019;25:382–394. doi: 10.1016/j.molmed.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: Small particle, big player. J Hematol Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2017;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 15.Lorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;7:848–856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tugay K, Guay C, Marques AC, Allagnat F, Locke JM, Harries LW, Rutter GA, Regazzi R. Role of microRNAs in the age-associated decline of pancreatic beta cell function in rat islets. Diabetologia. 2016;59:161–169. doi: 10.1007/s00125-015-3783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: Its role in tumour progression and response to therapy. Cancer Lett. 2014;356:321–331. doi: 10.1016/j.canlet.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 18.Pastushenko I, Blanpain C. EMT translation states during tumor progression and metastasis. Trends Cell Biol. 2018;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori G, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei SC, Yang J. Forcing through tumor metastasis: The interplay between tissue rigidity and epithelial-mesenchymal transition. Trends Cell Biol. 2016;26:111–120. doi: 10.1016/j.tcb.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu F, Zhang J, Hu G, Liu L, Liang W. Hypoxia and TGF-β1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell International. 2017;17:54. doi: 10.1186/s12935-017-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Xing T, Chen Y, Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed Pharmacother. 2018;106:1135–1143. doi: 10.1016/j.biopha.2018.07.042. [DOI] [PubMed] [Google Scholar]
- 23.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Jia M, Yuan K. MicroRNA-663b promotes cell proliferation and epithelial mesenchymal transition by directly targeting SMAD7 in nasopharyngeal carcinoma. Exp Ther Med. 2018;16:3129–3134. doi: 10.3892/etm.2018.6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YL, Shen Y, Xu JP, Han K, Zhou Y, Yang S, Yin JY, Min DL, Hu HY. Pterostilbene suppresses human endometrial cancer cells in vitro by down-regulating miR-663b. Acta Pharmacol Sin. 2017;38:1394–1400. doi: 10.1038/aps.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu Y, Ye W, Gu YL, Sun P. Blockade of miR-663b inhibits cell proliferation and induces apoptosis in osteosarcoma via regulating TP73 expression. Bratisl Lek Listy. 2018;119:41–46. doi: 10.4149/BLL_2018_009. [DOI] [PubMed] [Google Scholar]
- 27.Stanley P. Biological consequences of overexpressing or eliminating N-acetylglucosaminyltransferase-TIII in the mouse. Biochim Biophys Acta. 2002;1573:363–368. doi: 10.1016/S0304-4165(02)00404-X. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, Liu Y, Yu P, Qu J, Guo Y, Li W, Wang S, Zhang J. MiR-23a transcriptional activated by Runx2 increases metastatic potential of mouse hepatoma cell via directly targeting Mgat3. Sci Rep. 2018;8:7366. doi: 10.1038/s41598-018-25768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinho SS, Reis CA, Paredes J, Magalhães AM, Ferreira AC, Figueiredo J, Xiaogang W, Carneiro F, Gärtner F, Seruca R. The role of N-acetylglucosaminyltransferase III and V in the post-transcriptional modifications of E-cadherin. Hum Mol Genet. 2009;18:2599–2608. doi: 10.1093/hmg/ddp194. [DOI] [PubMed] [Google Scholar]
- 30.Kohler RS, Anugraham M, López MN, Xiao C, Schoetzau A, Hettich T, Schlotterbeck G, Fedier A, Jacob F, Heinzelmann-Schwarz V. Epigenetic activation of MGAT3 and corresponding bisecting GlcNAc shortens the survival of cancer patients. Oncotarget. 2016;7:51674–51686. doi: 10.18632/oncotarget.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimura M, Ihara Y, Matsuzawa Y, Taniguchi N. Aberrant glycosylation of E-cadherin enhances cell-cell binding to suppress metastasis. J Biol Chem. 2016;271:13811–13815. doi: 10.1074/jbc.271.23.13811. [DOI] [PubMed] [Google Scholar]
- 32.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichim TE, Zhong Z, Kaushal S, Zheng X, Ren X, Hao X, Joyce JA, Hanley HH, Riordan NH, Koropatnick J, et al. Exosomes as a tumor immune escape mechanism: Possible therapeutic implications. J Transl Med. 2008;6:37. doi: 10.1186/1479-5876-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. Virol. 2017;91:e02225–16. doi: 10.1128/JVI.02225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuzillet C, De Gramont A, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. Perspectives of TGF-β inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5:78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huynh LK, Hipolito CJ, Ten Dijke P. A perspective on the development of TGF-β inhibitors for cancer treatment. Biomolecculars. 2019;9:743. doi: 10.3390/biom9110743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park SJ, Choi YS, Lee S, Lee YJ, Hong S, Han S, Kim BC. BIX02189 inhibits TGF-β1-induced lung cancer cell metastasis by directly targeting TGF-β type I receptor. Cancer Lett. 2016;381:314–322. doi: 10.1016/j.canlet.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Lee J, You D, Jeong Y, Jeon M, Yu J, Kim SW, Nam SJ, Lee JE. Berberine suppresses cell motility through downregulation of TGF-β1 in triple negative breast cancer cells. Cell Physiol Biochem. 2018;45:795–807. doi: 10.1159/000487171. [DOI] [PubMed] [Google Scholar]
- 39.Yu F, Zhang X, Sun C, Xu W, Xia J. Downregulation of miRNA-663b protects against hypoxia-induced injury in cardiomyocytes by targeting BCL2L1. Exp Ther Med. 2020;19:3581–3588. doi: 10.3892/etm.2020.8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai H, An Y, Chen X, Sun D, Chen T, Peng Y, Zhu F, Jiang Y, He X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7:86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 42.Tan Z, Wang C, Li X, Guan F. Bisecting N-acetylglucosamine structures inhibit hypoxia-induced epithelial-mesenchymal transition in breast cancer cells. Front Physiol. 2018;9:210. doi: 10.3389/fphys.2018.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Wang Y, Qian Y, Wu X, Zhang Z, Liu X, Zhao R, Zhou L, Ruan Y, Xu J, et al. Discovery of specific metastasis-related N-glycan alterations in epithelial ovarian cancer based on quantitative glycomics. PLoS One. 2014;9:e87978. doi: 10.1371/journal.pone.0087978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miwa HE, Koba WR, Fine EJ, Giricz O, Kenny PA, Stanley P. Bisected, complex N-glycans and galectins in mouse mammary tumor progression and human breast cancer. Glycobiology. 2013;23:1477–1490. doi: 10.1093/glycob/cwt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akama R, Sato Y, Kariya Y, Isaji T, Fukuda T, Lu L, Taniguchi N, Ozawa M, Gu J. N-acetylglucosaminyltransferase III expression is regulated by cell-cell adhesion via the E-cadherin-catenin-actin complex. Proteomics. 2008;8:3221–3228. doi: 10.1002/pmic.200800038. [DOI] [PubMed] [Google Scholar]
- 46.Allam H, Johnson BP, Zhang M, Lu Z, Cannon MJ, Abbott KL. The glycosyltransferase GnT-III activates Notch signaling and drives stem cell expansion to promote the growth and invasion of ovarian cancer. J Biol Chem. 2017;292:16351–16359. doi: 10.1074/jbc.M117.783936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshimura M, Ihara Y, Ohnishi A, Ijuhin N, Nishiura T, Kanakura Y, Matsuzawa Y, Taniguchi N. Bisecting N-acetylglucosamine on K562 cells suppresses natural killer cytotoxicity and promotes spleen colonization. Cancer Res. 1996;56:412–418. [PubMed] [Google Scholar]
- 48.Yoshimura M, Ihara Y, Nishiura T, Okajima Y, Ogawa M, Yoshida H, Suzuki M, Yamamura K, Kanakura Y, Matsuzawa Y, Taniguchi N. Bisecting GlcNAc structure is implicated in suppression of stroma-dependent haemopoiesis in transgenic mice expressing N-acetylglucosaminyltransferase III. Biochem J. 1998;331:733–742. doi: 10.1042/bj3310733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagae M, Kizuka Y, Mihara E, Kitago Y, Hanashima S, Ito Y, Takagi J, Taniguchi N, Yamaguchi Y. Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V. Nat Commun. 2018;9:3380. doi: 10.1038/s41467-018-05931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taniguchi N, Kizuka Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res. 2015;126:11–51. doi: 10.1016/bs.acr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Xu J, Li L, Ianni A, Kumari P, Liu S, Sun P, Braun T, Tan X, Xiang R, Yue S. MGAT3-mediated glycosylation of tetraspanin CD82 at asparagine 157 suppresses ovarian cancer metastasis by inhibiting the integrin signaling pathway. Theranostics. 2020;10:6467–6482. doi: 10.7150/thno.43865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo C, Liu T, Zhang S, Guo K, Li M, Qin X, Liu Y. Reduced N-acetylglucosaminyltransferase III expression via Smad3 and Erk signaling in TGF-β1-induced HCC EMT model. Discov Med. 2017;23:7–17. [PubMed] [Google Scholar]
- 53.Xu Q, Isaji T, Lu Y, Gu W, Kondo M, Fukuda T, Du Y, Gu J. Roles of N-acetylglucosaminyltransferase III in epithelial-to-mesenchymal transition induced by transforming growth factor β1 (TGF-β1) in epithelial cell lines. J Biol Chem. 2012;287:16563–16574. doi: 10.1074/jbc.M111.262154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study can be obtained from the corresponding author upon reasonable request. The RNA sequencing data has been uploaded to the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with data number GSE163507.