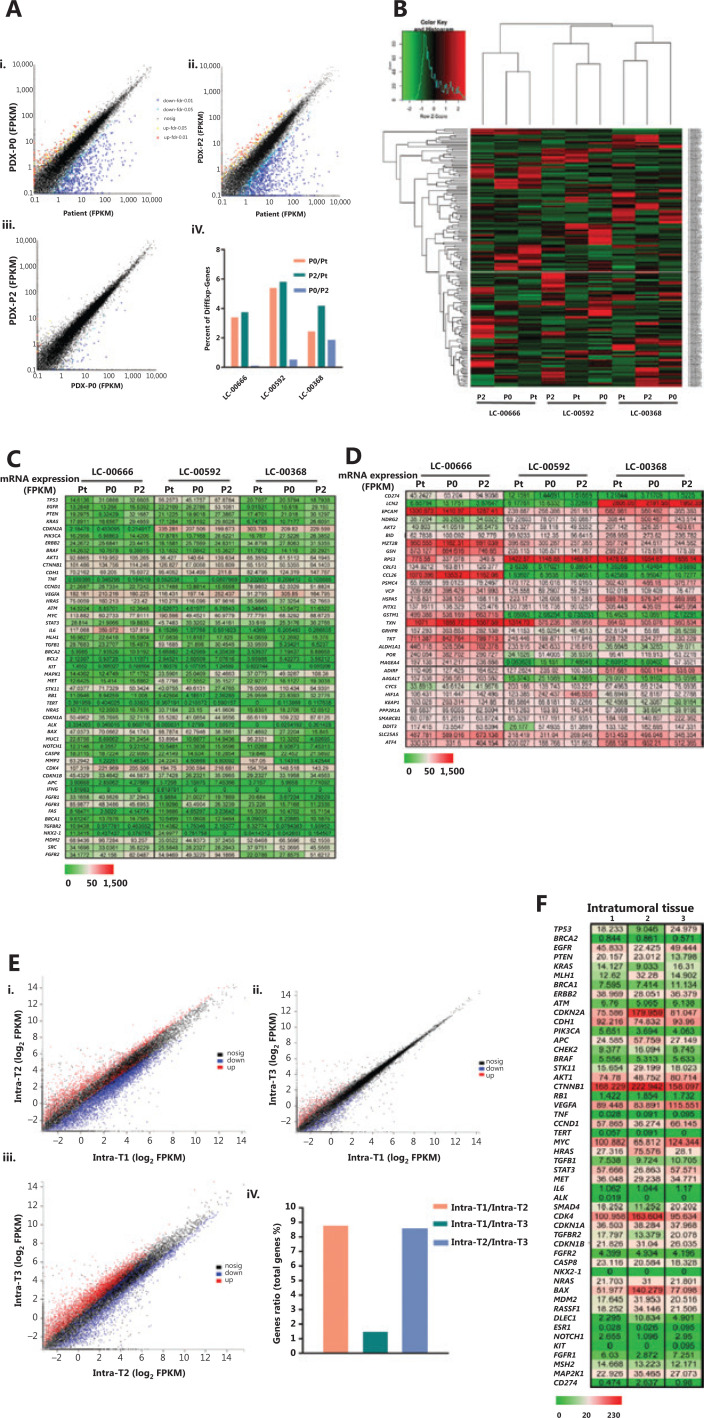
Figure 4.
Gene expression analysis between primary patient tumors and patient-derived xenographs (PDXs). (A) Comparison of genome-wide gene expression profiles between matched patient primary non-small cell lung cancer (NSCLC) tumor (patient LC-00666), the corresponding passage 0 (P0) and passage 2 (P2) of xenografts (i, ii, iii). Percent of differentially expressed genes in 3 PDX matched groups (LC-00666, LC-00592, and LC-00368) (iv). (B) Heat map of cancer-related gene expressions in 3 PDX matched groups. The dendrogram shows unsupervised hierarchical clustering of samples according to gene expression patterns. (C) Gene level expression (FPKM) values for the top 50 oncogenes related to NSCLC, and (D) Another 32 differentially expressed oncogenes are presented by the heat map (green = lower than the median, red = greater than the median). (E) Comparison of genome-wide gene expression profiles and (F) NSCLC-related gene expression (FPKM) values between three pieces of intratumoral tissues from one PDX (P4 of LC-00536).