Figure 1. Overview of short-read sequencing technologies.
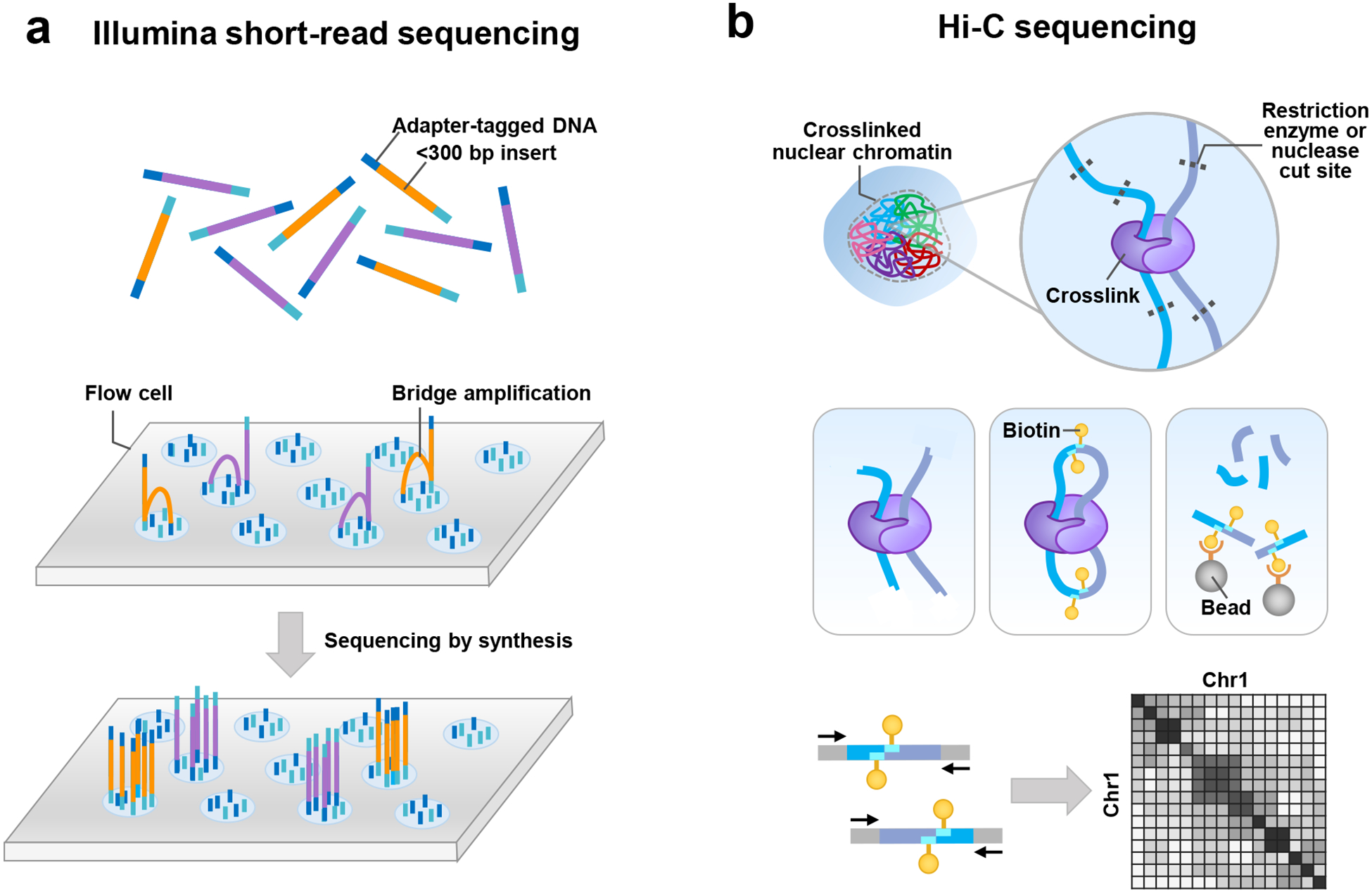
a) In short-read sequencing by Illumina, DNA fragments (yellow and purple) are ligated to adapters (blue and aqua) that contain unique molecular identifiers as well as sequences complementary to the oligonucleotides that are attached to the surface of a flow cell. The modified DNA is loaded onto a flow cell, and the adapters from the modified DNA hybridize to the oligonucleotides that coat the surface of the flow cell. Once the fragments have attached, cluster generation begins, where thousands of copies of each fragment are generated through a process known as bridge amplification. In this process, the strand folds over, and the adapter on the end of the molecule hybridizes to another oligonucleotide in the flow cell. A polymerase incorporates nucleotides to build double-stranded bridges of the DNA molecules, which are subsequently denatured to leave single-stranded DNA fragments tethered to the flow cell. This process is repeated over and over, generating several million dense clusters of double-stranded DNA. After bridge amplification, the reverse DNA strands are cleaved and washed away, leaving only the forward strands. Then, sequencing by synthesis begins, in which fluorescently labeled deoxyribonucleotide triphosphates (dNTPs) are incorporated into the newly synthesized DNA strand at each cycle. After incorporation, a laser excites the fluorophore on the strand, which emits a characteristic fluorescence emission signal that corresponds to the base. b) In Hi-C sequencing, nuclear chromatin is crosslinked with formaldehyde, which covalently bonds protein-DNA complexes in close proximity to each other. Crosslinked chromatin is digested with a restriction enzyme or nuclease, and single-stranded DNA overhangs are filled in and repaired with biotin-linked nucleotides before religating the DNA. Chemical crosslinks are reversed, proteins degraded, and the purified DNA is nonspecifically sheared (for example, by sonication). Biotin-labeled DNA is pulled down with streptavidin-conjugated beads and paired-end sequenced to reveal the junctions between two DNA loci (light and dark blue). Because the contact frequency between pairs of loci strongly correlates with distance, the majority of sequenced junctions encompass two loci from the same chromosome. As a result, Hi-C data can be used to provide linkage information between pairs of loci tens of megabases apart on a single chromosome (as shown in the contact map).
