Figure 2. Overview of long-read sequencing technologies.
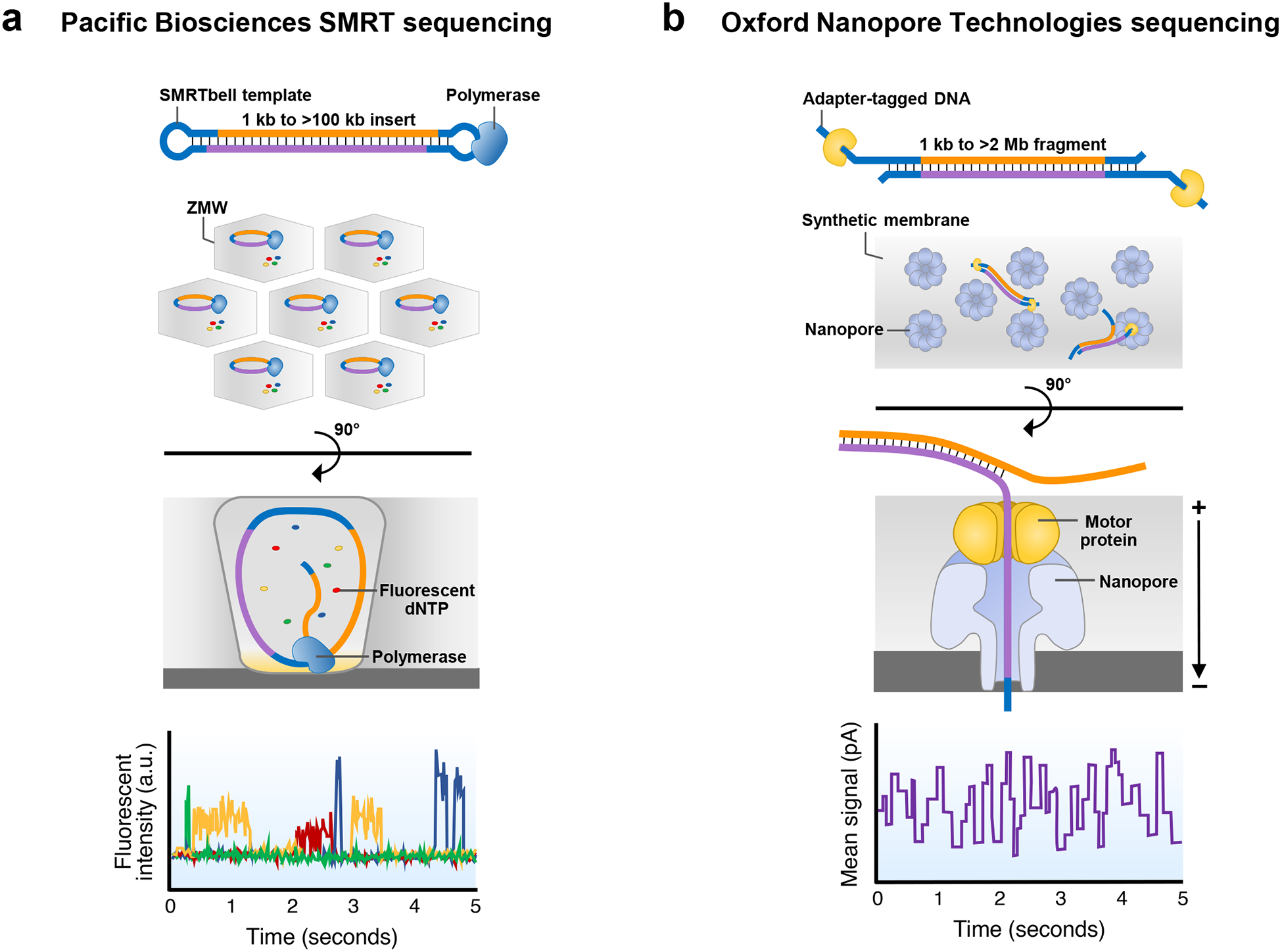
a) In single-molecule, real-time (SMRT) sequencing by Pacific Biosciences (PacBio), DNA (yellow for forward strand, dark blue for reverse strand) is fragmented and ligated to hairpin adapters (light blue) to form a topologically circular molecule known as a SMRTbell. Once the SMRTbell is generated, it is bound by a DNA polymerase and loaded onto a SMRT Cell for sequencing. Each SMRT Cell can contain up to eight million zero-mode waveguides (ZMWs), which are chambers that hold picoliter volumes. A light penetrates the lower 20–30 nm of each well, reducing the detection volume of the well to only 20 zeptoliters (10−21 liters). As the DNA mixture floods the ZMWs, the SMRTbell template and polymerase become immobilized on the bottom of the chamber. Fluorescently labeled dNTPs are added to begin the sequencing reaction. As the polymerase begins to synthesize the new strand of DNA, a fluorescent dNTP is briefly held in the detection volume, and a light pulse from the bottom of the well excites the fluorophore. Unincorporated dNTPs are not typically excited by this light but, in rare cases, can become excited if they diffuse into the excitation volume, thereby contributing to noise and error in PacBio sequencing. The light emitted from the excited fluorophore is detected by a camera, which records the wavelength and relative position of the incorporated base in the nascent strand. The phosphate-linked fluorophore is then cleaved from the nucleotide as part of the natural incorporation of the base into the new strand of DNA and released into the buffer, preventing fluorescent interference during the subsequent light pulse. The DNA sequence is determined by the changing fluorescence emissions that are recorded within each ZMW, with a different color corresponding to each DNA base (for example, green, T; yellow, C; red, G; blue, A). b) In Oxford Nanopore Technologies (ONT) sequencing, arbitrarily long DNA (orange for forward strand, purple for reverse strand) are tagged with sequencing adapters (light blue) preloaded with a motor protein on one or both ends. The DNA is combined with tethering proteins and loaded onto the flow cell for sequencing. The flow cell contains thousands of protein nanopores embedded in a synthetic membrane, and the tethering proteins bring the DNA molecules toward these nanopores. Then, the sequencing adapter inserts into the opening of the nanopore, and the motor protein begins to unwind the double-stranded DNA. An electric current is applied, which, in concert with the motor protein, drives the negatively charged DNA through the pore at a rate of about 450 bases per second. As the DNA moves through the pore, it causes characteristic disruptions to the current, known as a ‘squiggle’ [G]. Changes in current within the pore correspond to a particular k-mer (i.e., a string of DNA bases of length k) which is used to identify the DNA sequence.
