Figure 5. Long-read data provides insights into the biological relevance of structural variation and human evolution and diversity.
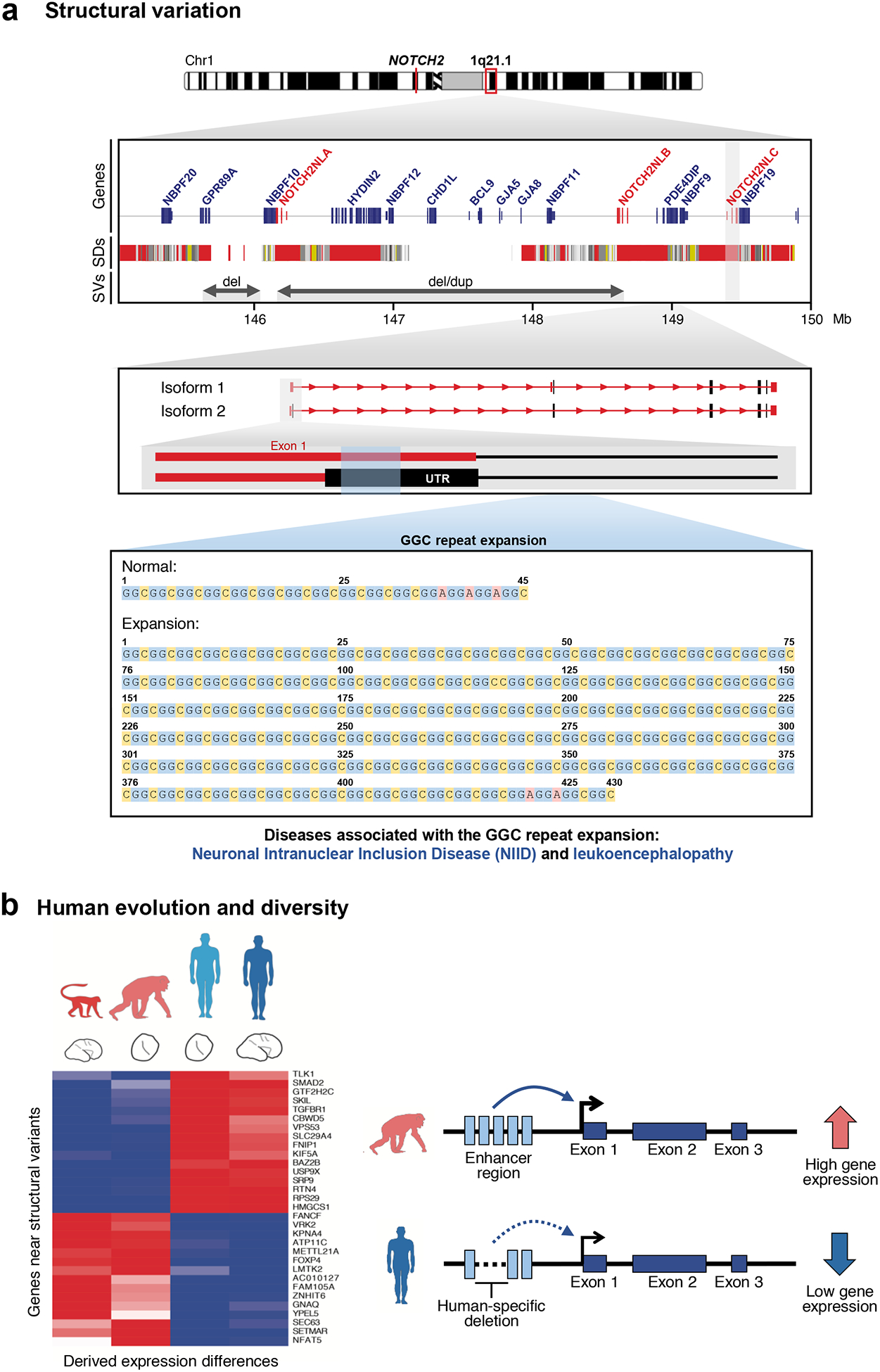
a) The NOTCH2NLA, B, and C genes are located within chromosome 1q21.1, a segmental duplication-rich region of the genome partially assembled by PacBio CLR sequencing of BAC clones116. The region was originally incorrectly assembled in the human reference genome116. Deletions and duplications mediated by the segmental duplication-rich region can cause thrombocytopenia-absent radius (TAR) syndrome165 as well as distal 1q21.1 deletion/duplication syndrome119,166. High-quality sequencing of the region allowed the breakpoints of these disease-causing rearrangements to be better defined and improved the annotation of human-specific NOTCH2NL duplicate genes116. Subsequent sequencing of patients affected with neuronal intranuclear inclusion disease (NIID) and leukoencephalopathy using long-read PacBio CLR and ONT sequencing recently identified a GGC repeat expansion in Exon 1 of NOTCH2NLC in affected patients66 (exons are in red; untranslated regions (UTRs) are in gray). Expansion of the repeat is associated with the production of anti-sense transcripts whose role is uncertain but may interfere with the expression and regulation of the gene family. Figure adapted from Ref. 66. SDs, segmental duplications; SVs, structural variants. b) Heatmap of differentially expressed genes located near structural variants in chimpanzee and human. Differences in macaque, chimpanzee, and human brain expression are shown for genes where a human-specific structural variant maps within 50 kbp of a transcription start and end. Structural changes, such as a deletion of an enhancer region as shown here, can cause changes in gene expression fundamental to brain development30.
