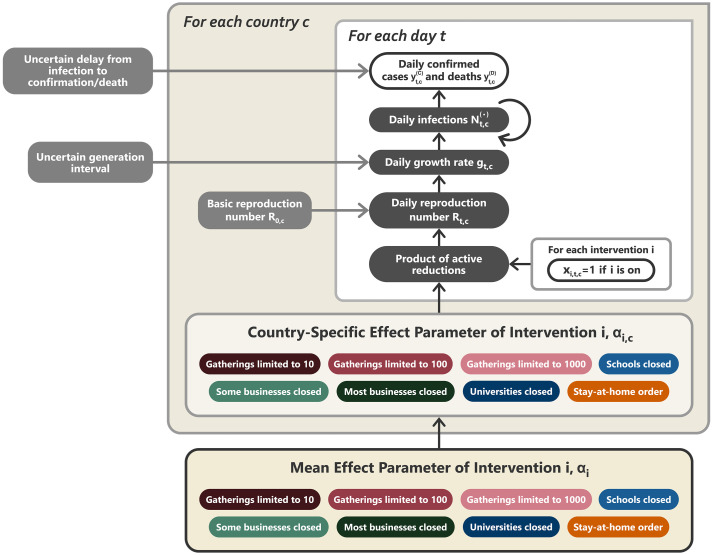
Fig. 5. Model overview.
Unshaded, white nodes are observed. From bottom to top: The mean effect parameter of NPI i is αi, and the country-specific effect parameter is αi,c. On each day t, a country’s daily reproduction number Rt,c depends on the country’s basic reproduction number R0,c and the active NPIs. The active NPIs are encoded by xi,t,c, which is 1 if NPI i is active in country c at time t, and 0 otherwise. Rt,c is transformed into the daily growth rate gt,c using the generation interval parameters and subsequently is used to compute the new infections and that will subsequently become confirmed cases and deaths, respectively. Finally, the expected numbers of daily confirmed cases and deaths are computed using discrete convolutions of with the relevant delay distributions. Our model uses both case and death data; it splits all nodes above the daily growth rate gt,c into separate branches for deaths and confirmed cases. We account for uncertainty in the generation interval, infection-to–case confirmation delay, and the infection-to-death delay by placing priors over the parameters of these distributions.