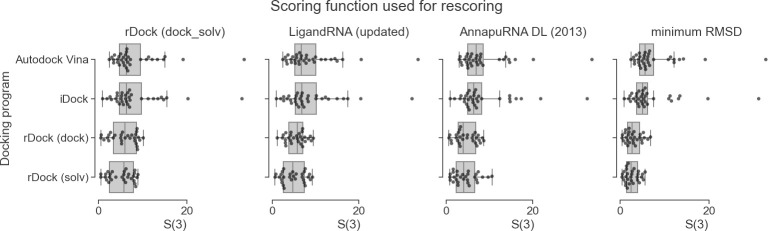
Fig 4. Comparison of the performance of three scoring functions (rDock dock_solv, LigandRNA, and AnnapuRNA) expressed as RMSD to the reference pose of best in top three scored poses (S(3)), calculated for four docking programs.
The fourth column represents the best (lowest) RMSD obtained during docking for each program. Each dot represents one complex from the testing set. Docking was performed with the native conformation of a ligand as an input.