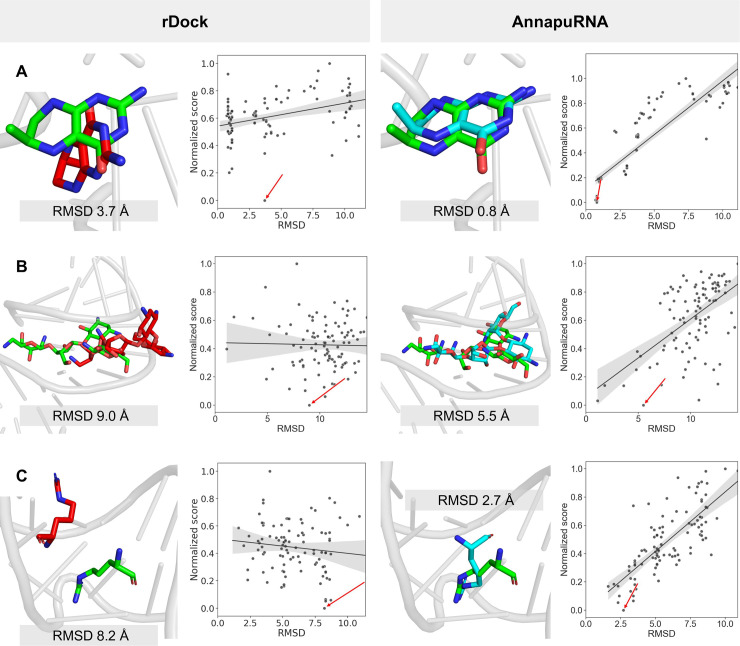
Fig 6. Selected docking solutions for structures from the testing set.
Best pose found by rDock (with dock desolvation potential, left) and AnnapuRNA (DL 2013, right), together with scatter plots of RMSD and normalized score. Scatter plots show a regression line with a 95% confidence, and the top scoring pose according to each method is indicated by a red arrow. (A) 3SUX (Crystal structure of THF riboswitch, bound with THF fragment), (B) 1FYP (Decoding region A-site in complex with Paromomycin) and (C) 1AJU (HIV-2 TAR-argininamide complex). RNA molecules are presented as a gray cartoon, ligands as sticks; reference structure—green, solution found by rDock—red, solution found by AnnapuRNA—cyan. Heteroatoms are colored: O—red, N—blue.