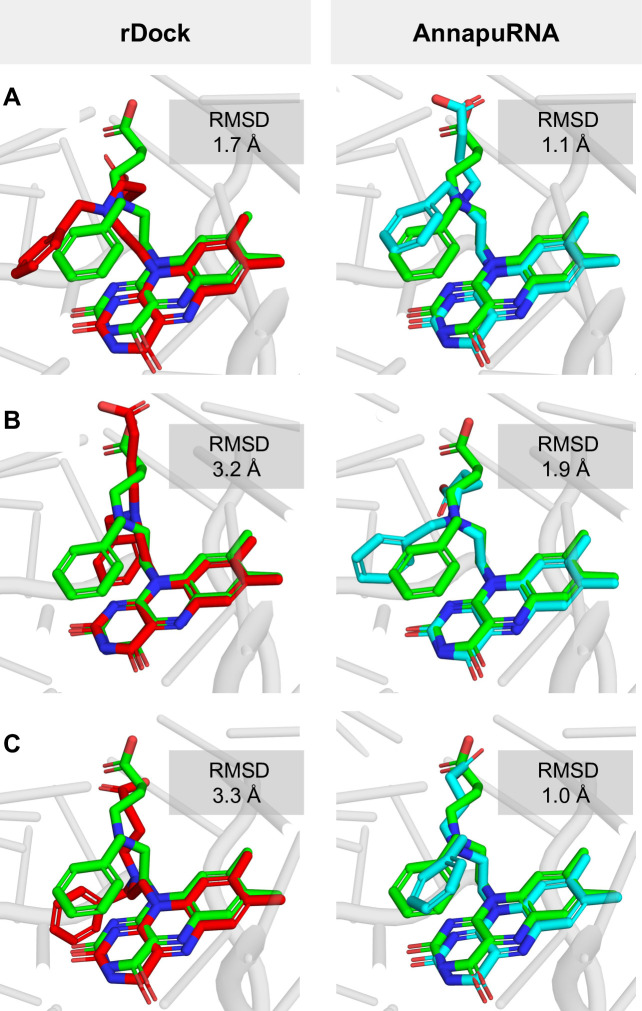
Fig 7. Prediction of the complex of FMN Riboswitch with 4-{benzyl[2-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)ethyl]amino}butanoic acid.
(A). Redocking experiment—ligand extracted from the crystal structure 6DN2 was redocked to this structure; (B). Ligand extracted from the crystal structure 6DN2 docked to FMN structure solved with a different ligand (2YIE); (C). Ligand extracted from the crystal structure 6DN2 docked to the low-resolution APO FMN structure (2YIF). All RNA structures are superimposed. The reference ligands structures are shown in green. Structures predicted by rDock (dock_solv) are shown in magenta (left), predicted by AnnapuRNA (DL 2013) in light blue (right).