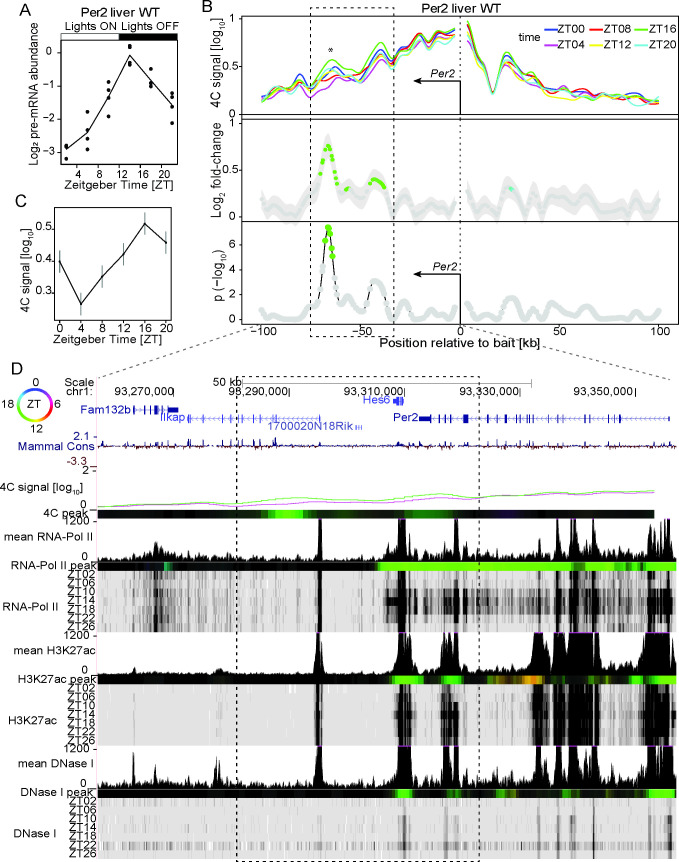
Fig 2. Time-resolved 4C-seq experiments revealed 24h rhythms in chromatin interactions of the Period2 promoter in WT mouse liver.
(A) Period2 pre-mRNA expression over time in WT mouse liver [47]. (B) 4C-seq signal over time from the Period2 TSS bait in WT mouse liver (top panel) and log2 fold change (middle panel) and −log10(p) from rhythmicity analyses [23]. Fragments with p< 0.01 are colored according to peak time in contact frequency (color-coding as in following top left circle panel D). * = local maximum in differential genomic contact. n=2. Dashed rectangle: region of rhythmic interaction. (C) 4C-seq signal over time adjacent to * (B). (D) 4C-seq signal at ZT04 (purple) and ZT16 (green), and time-resolved ChIP-seq signal for PolII, H3K27ac and DNase1 hypersensitivity in WT mouse liver [8]. Colored tracks represent peak time for the 4C-seq signal and chromatin marks, (top left circle for peak-time color code, Material and Methods). Dashed rectangle: region of rhythmic interaction. The region interacting with the promoter of Period2 at ZT16 coincided with multiple localized rhythmic signals in H3K27ac and DNase1 hypersensitivity peaking at ZT16. See S7B Fig for ChIP-seq signals of CTCF and core clock factors at the connected genomic regions.