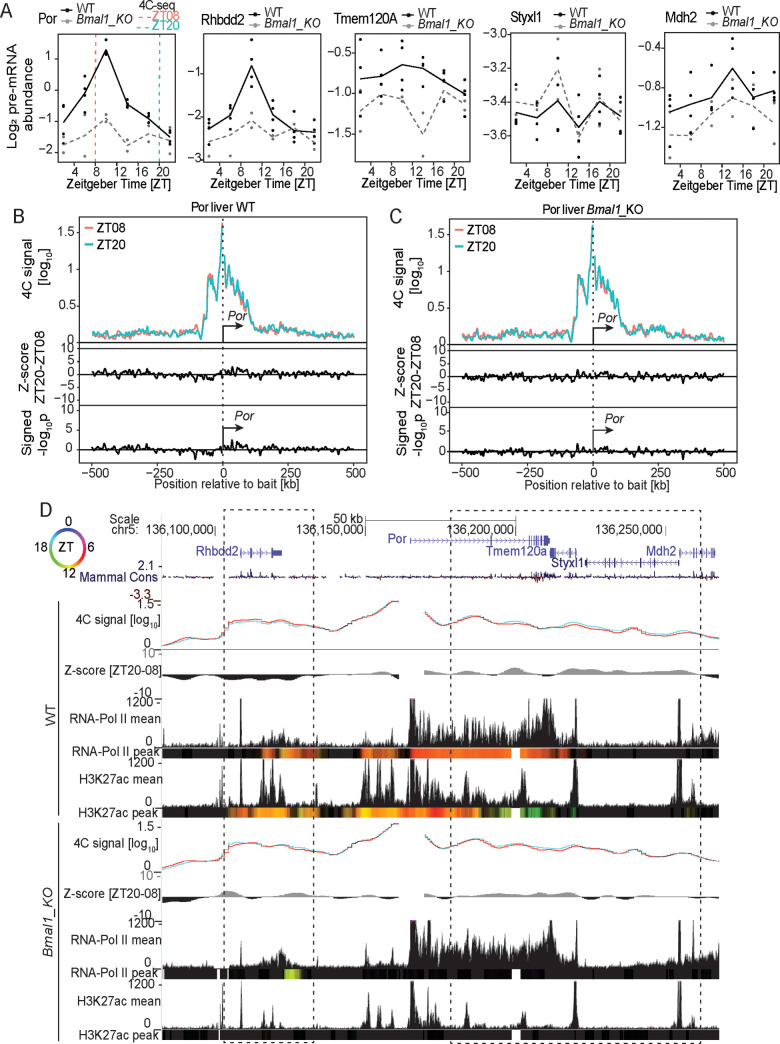
Fig 5. The Por gene stably connects surrounding genes having synchronized transcriptional dynamics in WT and Bmal1 KO livers.
(A) Por, Rhbdd2, Tmem120a, Styxl1 and Mdh2 pre-mRNAs accumulation over time in WT (solid line) and Bmal1 KO (dashed line) livers [47]. (B and C) 4C-seq signal from Por TSS bait at ZT08 (red) and ZT20 (green) in a genomic window of 1Mb surrounding the bait in livers of WT (B, n=3 at ZT08 and n=4 at ZT20) and Bmal1 KO (C, n=3) animals and the corresponding Z-scores and p-values. The most prominent interacting regions are located in the vicinity of Por. (D) Genome browser view of the Por 4C-seq signal at ZT08 (red line) and ZT20 (blue line) and the corresponding Z-scores in livers of WT and Bmal1 KO animals. Mean and peak time of PolII and H3K27ac ChIP-seq signals are shown for both the WT and Bmal1 KO conditions [8]. Colored tracks represent peak time in chromatin mark (color code in the top left circle, Material and Methods). Dashed rectangle: region of highest interaction frequency. PolII loading and H3K27ac oscillate in WT livers at interacting regions, notably at Por, Rhbdd2, and Tmem120a genes and peak around ZT10, consistently with the rhythmic transcription of these genes. The dynamics of chromatin marks is lost over the entire locus in arrhythmic Bmal1 KO livers, while 4C-seq signals are similar to WT condition. See S9B Fig for ChIP-seq signals of CTCF and core clock factors at the connected genomic regions.