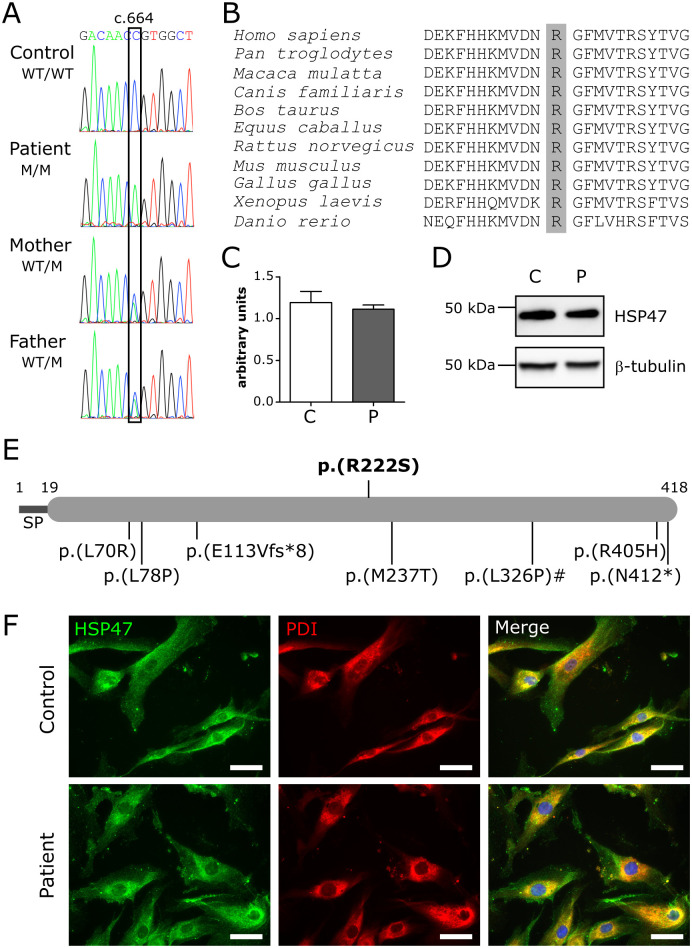
Fig 2. Variant identification and characterization.
(A) Electropherograms of the identified variant in the patient, unaffected parents and control. WT: wild-type, M: mutant. (B) Protein sequence alignment of different species. The affected p.Arg222 residue is shaded in grey. (C) RT-qPCR expression analysis for SERPINH1. Data are expressed as mean ± SEM. (D) Western blot analysis for HSP47. (E) HSP47 domain structure with inclusion of all pathogenic variants identified to date. The variant reported here is depicted above the structure. Numbers above the structure indicate amino acid residues. The pathogenic variant with a hashtag (#) depicts the only canine variant identified so far. The reported upstream genomic deletion is not depicted in this image. SP: signal peptide. (F) Immunocytochemical analysis showing colocalization of wild-type and R222S HSP47 with the ER-marker PDI. Nuclei are counterstained with DAPI. Scale bar = 50 μm. C: control and P: patient.