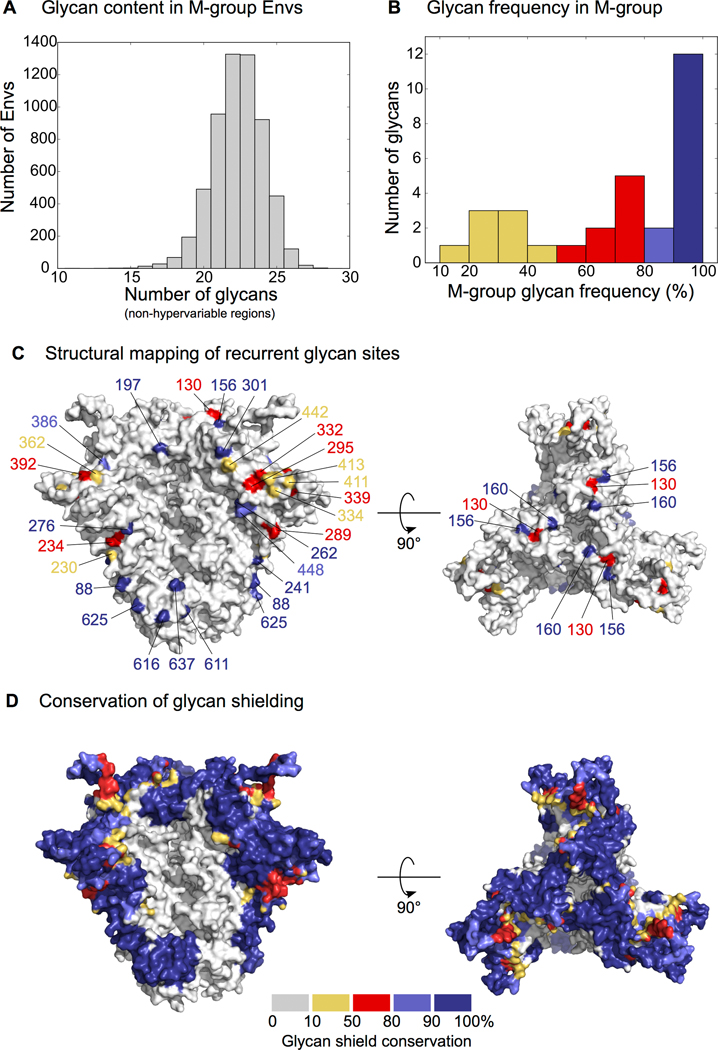
Figure 1.
Conservation of glycans and the glycan shield in M-group Envs.
(A) Distribution of unique (i.e. per protomer) glycans in an alignment of ~4,800 M-group Envs (2018 Filtered Web alignment from the Los Alamos HIV Database, www.hiv.lanl.gov). Hypervariable loop glycans are excluded. (B) Distribution of glycan frequency in M-group Envs. Hypervariable loop glycans and glycans with <10% frequency are excluded. (C) Structural mapping of glycan sites in (B). PDB: 5FYJ [1]. (D) Conservation of glycan shielding in M-group using the calculations from Wagh et al. [5*].
Original.