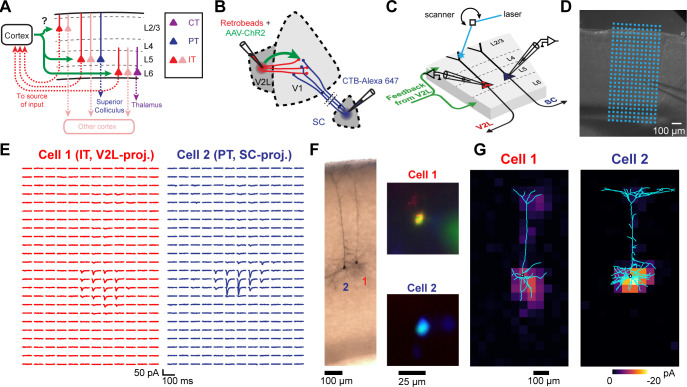
Figure 2. Measuring the strength and dendritic distribution of cortico-cortical (CC) inputs to different projection neurons.
(A) We probed the strength of CC inputs to looped and non-looped neurons in different cortical layers. (B) Example experiment configuration. Retrograde tracers are injected in two areas to label different projection neurons. One cortical area is also co-injected with adeno-associated virus (AAV)-channelrhodopsin-2 (ChR2) to express ChR2 in a specific CC projection. (C) Example of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) experiment. Pairs of neighboring retrogradely labeled neurons in the same cortical layer were sequentially recorded. During each recording, a laser beam was scanned over the dendrites of the cell at different locations in a grid pattern. (D) Brightfield image of an acute coronal cortical slice showing the recording pipette and photostimulation grid. (E) Excitatory postsynaptic currents (EPSCs) recorded from a pair of neighboring L5 neurons, evoked by photostimulating ChR2+ V2L→V1 FB terminals on a grid. (F) Left, dendritic morphology staining of the recorded pair. Right, identity of the recorded projection neuron was confirmed by fluorescence in the soma of both a retrograde tracer and a different-colored dye introduced from the internal patch pipette solution. (G) sCRACM maps of the recorded pair overlaid on their reconstructed dendrites. Responsive locations are color-coded to represent mean amplitude.