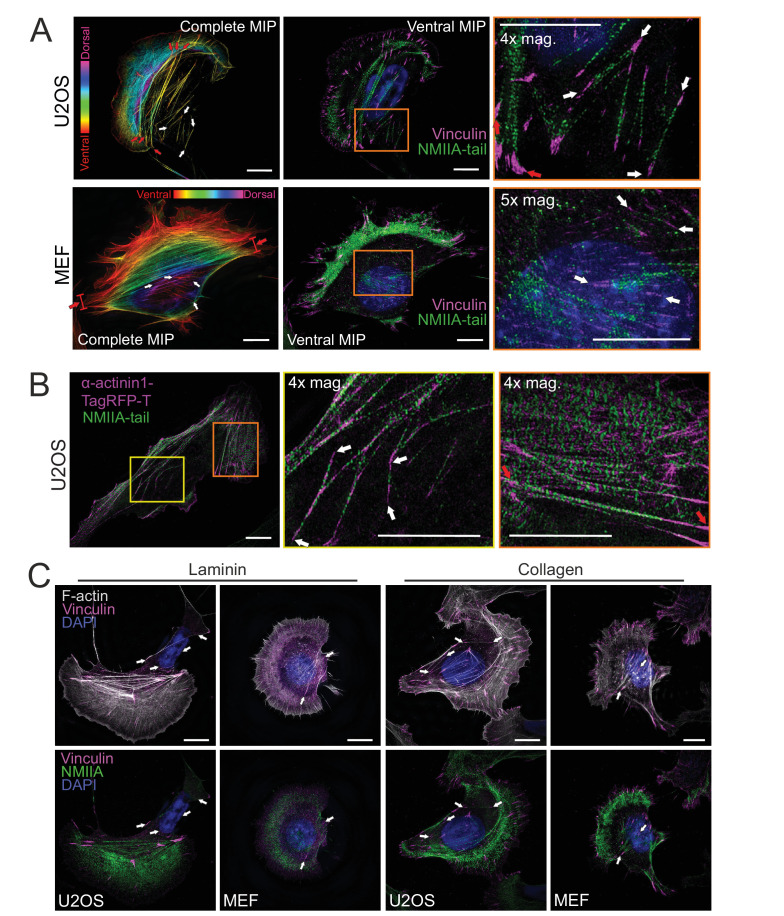
Figure 1. Stress fiber architecture of migrating cells.
3D-structured illumination microscopy (SIM) maximum intensity projections (MIPs) of the actomyosin networks in cells migrating on fibronectin. (A) Human osteosarcoma (U2OS) and mouse embryonic fibroblast (MEF) cells, where the panels on the left display complete MIPs. The panels in the middle show only the filament structures close to the ventral plane (‘ventral MIP’), and the panels on the right are magnifications of the boxed regions from the middle panels. Red arrows highlight ventral stress fibers, and white arrows indicate examples of thin cortical stress fibers that are embedded at the cell cortex. DAPI (blue) and phalloidin (gray) were applied to mark the F-actin and nucleus, respectively. Vinculin (magenta) and non-muscle myosin II (NMII)A tails (green) were detected with respective specific antibodies. (B) 3D-SIM MIP projection from the ventral plane of a U2OS cell transfected with mApple-NMIIA construct (motor, green) and stained with NMIIA-tail specific antibody (magenta, tail) and phalloidin to visualize F-actin. 4× and 10× magnifications (orange box and yellow dotted box, respectively) show the bipolar NMIIA filaments in cortical stress fibers. (C) Traction force microscopy (TFM) analysis of the forces exerted by ventral stress fibers (red arrows) and cortical stress fibers (white arrows) to the underlying substrate. On the left, exemplary image of a LifeAct-mKate1.31-expressing U2OS cell and the obtained force map with root mean square tractions (RMS). Quantification of the RMS forces between the two stress fiber subtypes (n = 41 for ventral stress fibers [SF] and 45 for cortical stress fibers) is shown on right as half-violin plot including binned individual data points and mean with ±1 standard deviation (SD). p = 1.56 × 10−12 (Mann–Whitney U-test, including outliers). Scale bars 10 µm and 5 µm for whole cell images and magnifications, respectively.