Fig. 2. Rates of putative loss-of-function variants (stop gained) relative to synonymous single nucleotide polymorphisms reported in recent whole genome re-sequencing projects (Table 1).
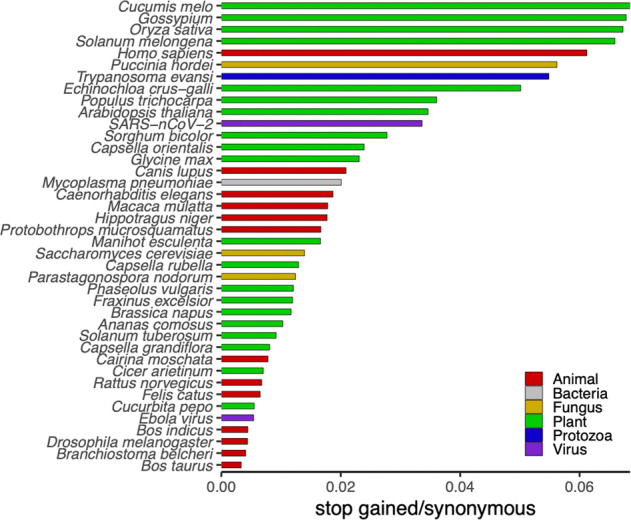
Species exhibit considerable differences in the ratio of stop gained to synonymous single nucleotide polymorphisms, with a 20-fold difference between the greatest (Cucumis melo) and fewest (Bos taurus) observations. The causes for these differences between species and, more generally, the (mal)adaptive nature of this extensive loss-of-function variation remain largely unknown.
