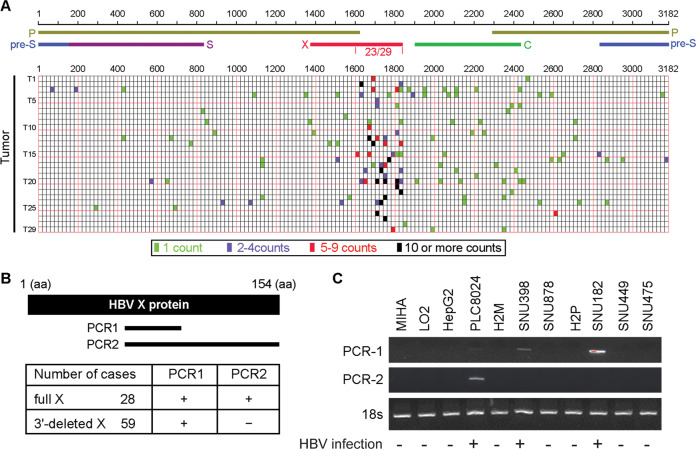
Fig. 1. HBV breakpoints within the HBV genome were clustered at 3′-end X gene region in HCC samples.
A The frequency of integration breakpoints within the HBV genome (NC_003977) is indicated by bars in 29 cases of tumor samples. Integration sites with different supporting counts are indicated by different colors. Genes encoding HBV pre-surface (blue), surface (purple), polymerase (yellow), core (green), and X (red) protein are also shown. B Two pairs of primers are designed for the detection of full-length and 3′-end deleted X gene. 66F + 100R flanks 360 nucleotides (PCR1), 66F + 154R flanks 462 nucleotides (PCR2). Expression of full-length and 3′-end deleted X gene in 87 HBV-related HCC samples were detected by qRT-PCR. C Expression pattern of both full-length and 3′-end deleted X gene in a panel of hepatoma cell lines was detected by RT-PCR, 18 s served as the internal reference.