Figure 4. SKIP Self-Association Mediated by the Middle Region Enhances Interaction with Kinesin-1.
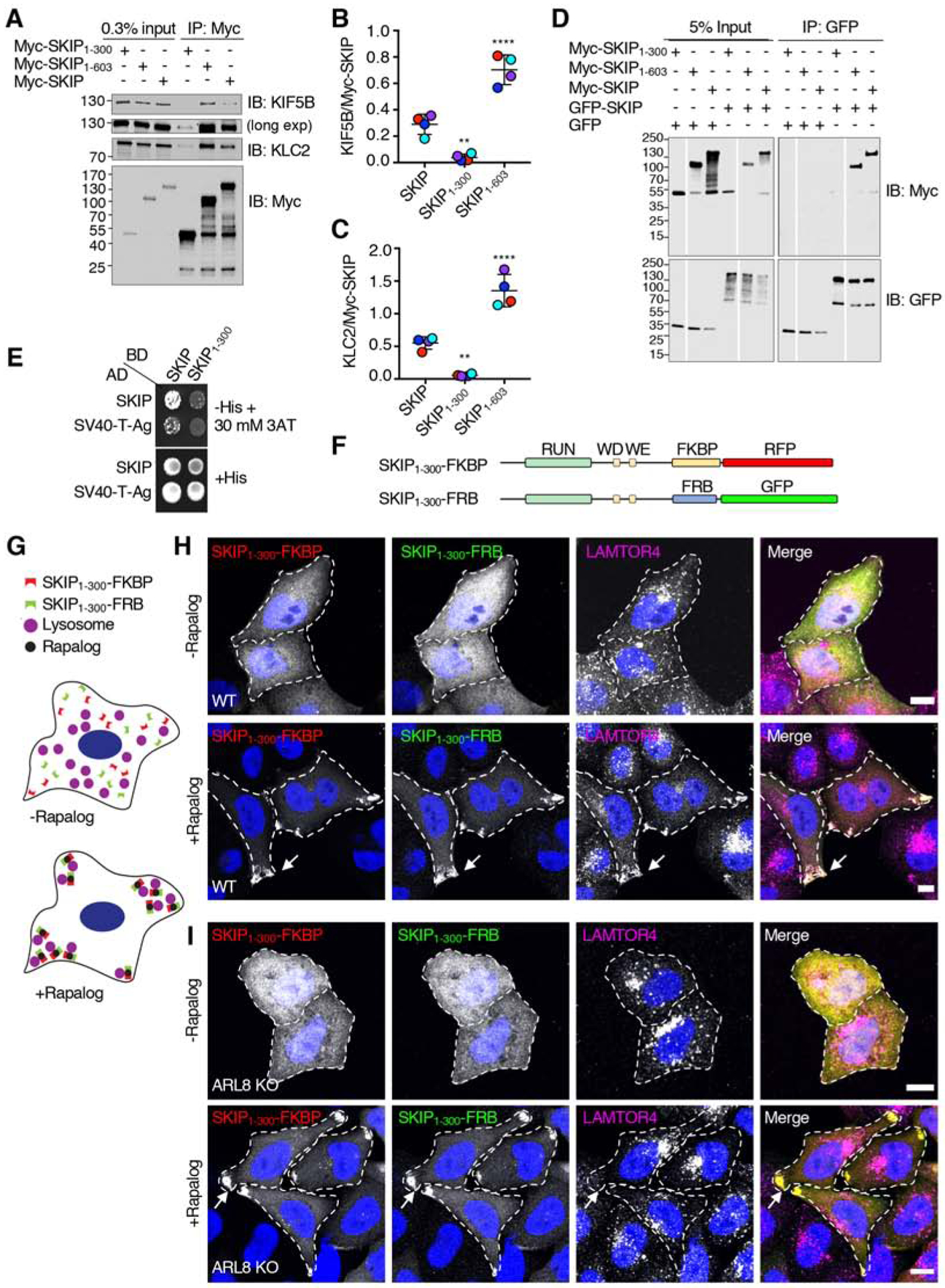
(A) Co-immunoprecipitation of Myc-SKIP constructs with endogenous kinesin-1. HEK293T cells were transfected with plasmids encoding the indicated Myc-SKIP constructs, subjected to immunoprecipitation with antibody to the Myc epitope and analyzed by SDS-PAGE and immunoblotting for the Myc epitope and the endogenous KIF5B and KLC2 subunits of kinesin-1. Immunoblotting for KIF5B is shown in short and long exposures.
(B,C) Quantification of experiments shown in panel A. Signal intensity was quantified using Image J. The ratio of KIF5B (B) and KLC2 (C) to immunoprecipitated Myc-SKIP was calculated. Values are the mean ± SD from 4 independent experiments. Statistical significance was calculated by one-way ANOVA followed by multiple comparisons to full-length SKIP using Dunnett’s test., **P<0.01, ****P<0.0001. Independent experiments are color coded.
(D) Co-immunoprecipitation of SKIP constructs. HEK293T cells were co-transfected with plasmids encoding the indicated Myc-SKIP constructs and full-length GFP-SKIP or GFP (control), subjected to immunoprecipitation with antibody to GFP, and analyzed by SDS-PAGE and immunoblotting with antibodies to the Myc epitope and GFP. Data are representative of 2 independent experiments with similar results. Vertical white lines indicate places where blots were cut to remove irrelevant lanes. All lanes are from the same blots in the same experiment. In A and D, numbers on the left indicate the positions of molecular mass markers (in kDa).
(E) Yeast two-hybrid analysis of the interaction of full-length SKIP or the SV40 T-antigen (control) fused to the β-Gal activation domain (AD), and full-length SKIP or SKIP1–300 fused to the β-Gal DNA-binding domain (BD). Growth in the absence of histidine (-His) indicates binding between the proteins. Thirty mM 3AT was added to reduce non-specific interactions. Data are representative of 3 experiments with similar results.
(F) Schematic representation of the constructs used to perform rapalog-induced dimerization of SKIP1–300 as previously described for microtubule motors [61]. Domains of SKIP are as described in the legend to Figure 1. SKIP constructs were C-terminally appended with FKBP-RFP or FRB-GFP.
(G) Cartoon representing the hypothetical effect of rapalog addition to WT HeLa cells co-expressing the constructs shown in panel F if dimerization of SKIP1–300 enables coupling of lysosomes to kinesin-1. SKIP1–300 plasmids tagged with FKBP-RFP or FRB-GFP are cytosolic in the absence of rapalog. Addition of rapalog induces the dimerization of FKBP with FRB, promoting the accumulation of SKIP1–300 with lysosomes at cell vertices.
(H) Confocal immunofluorescence microscopy of WT HeLa cells co-transfected with plasmids encoding the constructs shown in panel F, without (top panel) and with rapalog (bottom panel), and immunostained with antibody to the lysosomal marker LAMTOR4 (magenta channel). SKIP1–300-FKBP-RFP and SKIP1–300-FRB-GFP fluorescence is shown in the red and green channel, respectively. Nuclei were stained with DAPI (blue). Single channels are shown in grayscale with nuclei in blue. Cell edges are outlined. Arrows point to accumulation of SKIP1–300 constructs and LAMTOR4 at cell vertices. Scale bars: 10 μm.
(I) Same as panel H but using ARL8-KO HeLa cells.
The related Figure S2 shows the amino-acid sequence conservation of the SKIP middle region in different animal species.
