Figure 5. Shifting the balance of SRSF10 transcripts by transfecting either anti-short (S→L) and anti-long (L→S) SSÓs into HeLa cells.
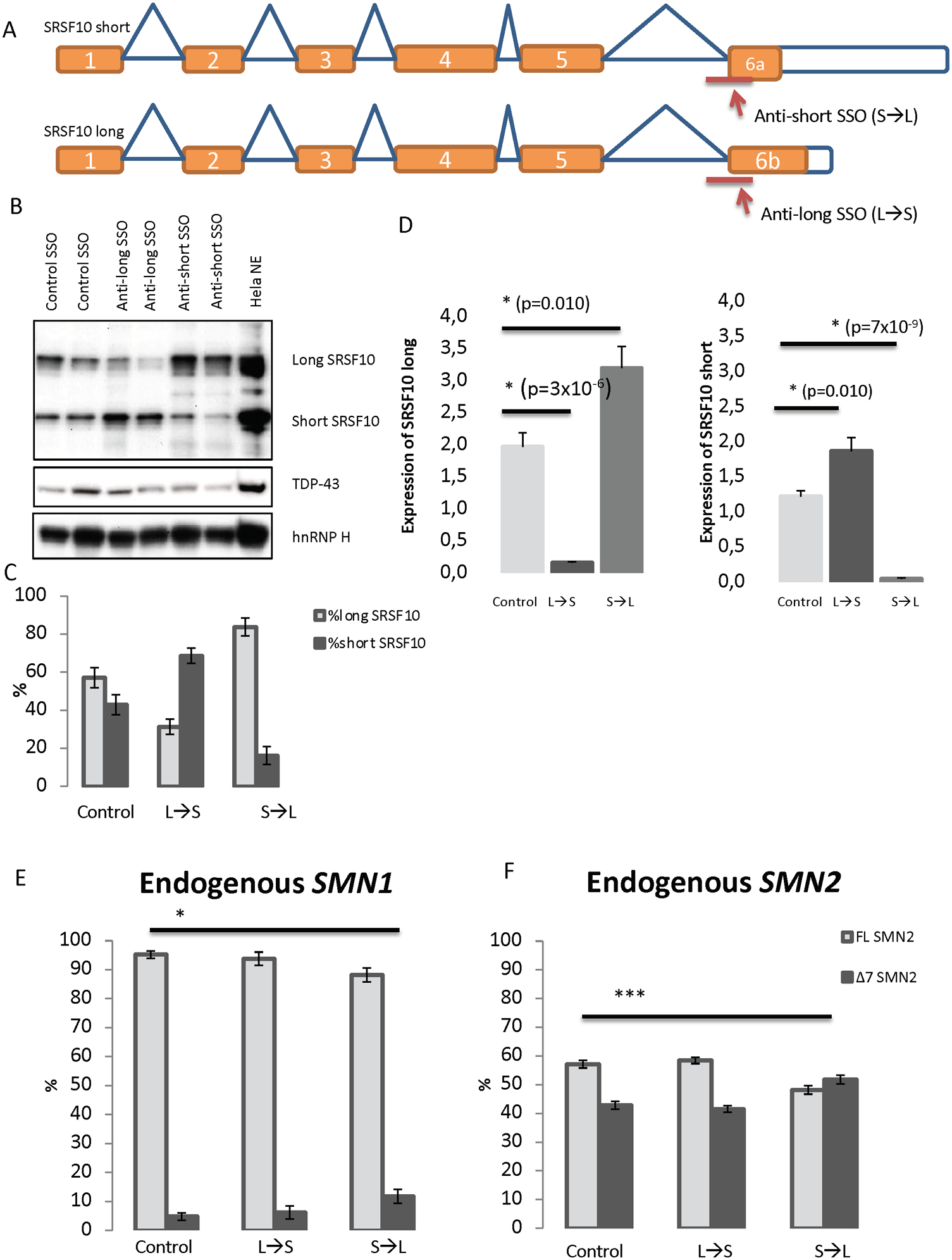
(A) SSOs were designed to target the 3śplice site of either exon 6a or exon 6b. This should alter the splicing of SRSF10 transcripts to either produce the long isoform (when treated with the anti-short SSO) or produce the short isoform (when treated with the anti-long SSO). (B) Validation of the shifted splicing pattern by Western blot analysis using antibody against SRSF10. Antibodies against TDP-43 and hnRNP H were used for controls. HeLa nuclear extract was included as a positive control. The blot displayed is representative of all replicates. (C) Densitometry quantification using ImageJ of replicates of Western blots. N=4. Error bars are displayed as SD/sqrt(n). (D) QPCR analysis with specific primers to detect the expression of SRSF10 long and SRSF10 short in SSO-treated samples. Expression was normalized to TBP. Errors bars are displayed as SD/sqrt(n). N>6. (E-F) HeLa cells were reverse-transfected with 40 nM anti-short, anti-long or control SSO for 48 h. The RNA was isolated and used for cDNA synthesis. PCR with primers in exons 6 and 8 of SMN was used. The PCR products were digested with DdeI to distinguish between PCR product originated from either SMN1 or SMN2. The digested products were quantified with an Agilent 2100 Bioanalyzer instrument. (*) indicates p<0.001 when performing a two-tailed Student T test. Error bars are displayed as STD. N=6.
