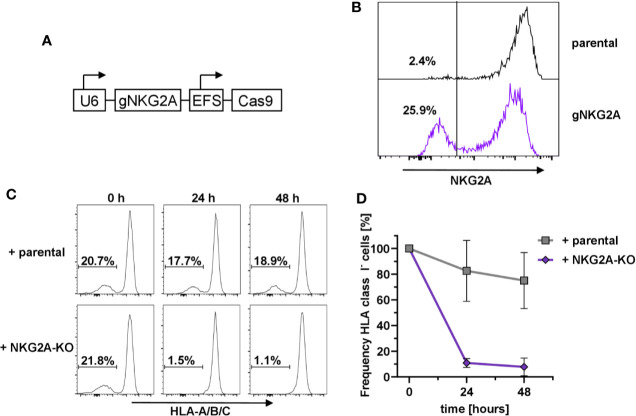
Figure 2.
Dependency on NKG2A expression for sc-HLA-E-mediated protection from NK cell fratricide (A) Outline of the lentiviral expression cassettes used for CRISPR/Cas9 mediated NKG2A-knockout. (B) Transduction with Cas9 and gNKG2A led to abrogation of NKG2A surface expression in primary NK cells 7 days post transduction. (C) Representative histograms showing the frequency of sc-HLA-E/B2M-KO NK cells in presence of B2M-competent sc-HLA-E NK cells upon addition of parental cells (upper panel) or NKG2A-KO NK cells (lower panel) at different time points. (D) Quantification of changes in sc-HLA-E/B2M-KO NK cell frequencies relative to the time point (0 h) when parental or NKG2A-KO NK cells were added. Statistical analysis was performed by two-way ANOVA with Geisser-Greenhouse correction (ε = 0.6972) for repeated measures and Holm-Sidak testing for multiple comparisons between each time point within both groups. For the samples containing NKG2A-KO NK cells, the decrease in frequency of HLA class I− was statistically significant at both time points (mean ± SD; n = 4).