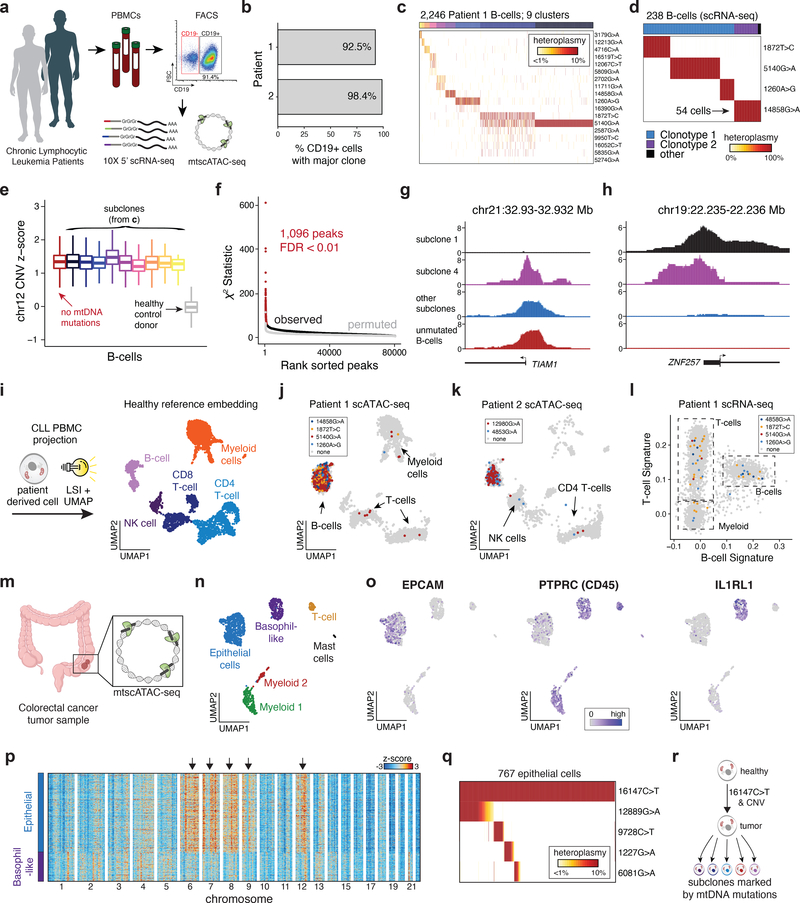
Figure 4 -. Clonal and functional heterogeneity in human malignancies resolved by somatic mtDNA mutations.
(a) Schematic of experimental design. Populations of peripheral blood mononuclear cells (PBMCs) from two CLL patients were separated by FACS or magnetic bead enrichment and profiled with mtscATAC-seq and 10× 5’ scRNA-seq. (b) Fraction of CD19+ cells with major B cell receptor (BCR) clonotype as determined from V(D)J receptor sequencing. (c) Inference of subclonal structure from somatic mtDNA mutations for patient 1. Cells (columns) are clustered based on mitochondrial genotypes (rows). Colors at the top of the heatmap represent clusters or putative subclones. Color bar, heteroplasmy (% allele frequency). (d) Clonotype receptors (columns) associated with somatic mtDNA mutations (rows) from patient 1. Colors at the top of the heatmap represent BCR clonotypes. Color bar, heteroplasmy (% allele frequency). (e) Estimated copy number of chromosome 12 across putative subclones for patient 1. Patient derived cells showed elevated DNA read counts of chromosome 12, consistent with a trisomy for this chromosome (Methods). Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. (f) Sub-clone associations with accessible chromatin. Red dots denote peaks associated at a false-discovery rate of <0.01. (g,h) Examples of subclone-associated differential accessibility peaks near the (g) TIAM1 and (h) ZNF257 promoters. (i) Schematic of scATAC projection framework using latent semantic indexing (LSI) and UMAP. A healthy PBMC reference embedding with indicated cell types is shown. (j) Projection of cells collected from Patient 1 and (k) Patient 2. Colors indicate cells positive for indicated somatic mtDNA mutations. Non-B-cells are highlighted. (l) Gene signature plots of PBMCs from single-cell RNA-seq for Patient 1 corroborating mtDNA mutations in non-B-cells. (m) Schematic showing mtscATAC-seq profiling of a colorectal cancer resection specimen. (n) Two dimensional embedding of all quality controlled tumor derived cells using UMAP showing the distribution of cells based on Louvain clustering and annotation based on marker gene scores as exemplified in panel (o) and Extended Data Fig. 4l. (o) Projection of marker gene scores for indicated genes EPCAM, PTPRC and IL1RL1. Color bar, gene score activity. (p) Inferred CNV profiles for indicated cell types (x axis) and chromosomes. Arrows indicate relative increase of copy numbers in the epithelial tumor cells. Cells from the basophil-like population are shown as a control group of cells. Color bar, z-score transformed fragment abundance. (q) Inference of subclonal structure from somatic mtDNA mutations in colorectal cancer. Epithelial cells (columns) are clustered based on mitochondrial genotypes (rows). Color bar, heteroplasmy (% allele frequency). (r) Putative model of clonal evolution of the profiled colorectal cancer specimen as suggested based on integrated analysis of nuclear CNV and somatic mtDNA mutation profiles.