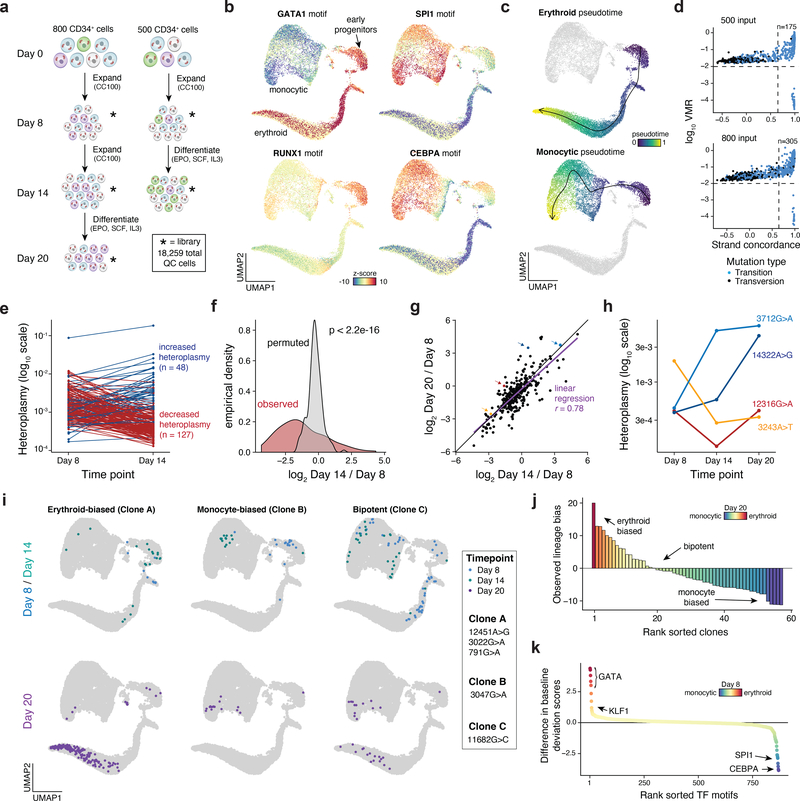
Figure 5 -. Clonal lineage tracing across accessible chromatin landscapes and time in an in vitro model of hematopoiesis.
(a) Schematic of experimental design. Approximately 800 or 500 CD34+ HSPCs were derived from the same donor, expanded, and differentiated in two independent cultures over the course of 20 days as shown. Stars represent timepoints/ populations of cells that were profiled via mtscATAC-seq. (b) Two dimensional embedding of all quality controlled cells using UMAP. Single-cell TF motif deviation scores for indicated factors are shown in color for all cells. (c) Pseudotime trajectories for monocytic and erythroid trajectories are depicted. (d) Identification of high confidence variants derived from both cultures. The number of variants passing both thresholds (dotted lines) is indicated. (e) Changes in heteroplasmy for 175 variants identified from the 500 input culture from day 8 to day 14. Values represent the mean over all single-cells in the library. (f) Increased variability in heteroplasmy shifts for the 500 cell input culture. P-value is reported from a two-sided Kolmogorov–Smirnov test comparing the observed and permuted distributions log fold-changes of heteroplasmy. (g) Comparison of heteroplasmy shifts for the 800 cell input culture. Linear regression indicates that most of the variability in heteroplasmy changes at the late time point (day 20, y-axis) can be explained by the intermediate time point (day 14, x-axis). Colored dots are mutations highlighted in the next panel. (h) Heteroplasmy trajectories for four selected mutations from (g). Values represent the mean over all single-cells in the library for the indicated time point. (i) Three examples of clonal populations marked by indicated mutations identified in the 800 cell input culture that result in erythroid, monocytic, or bipotent lineage outcomes. (j) Systematic identification of clonal outcomes using the late time point (day 20). y-axis depicts the difference between z-score in erythroid and monocytic bias of a single clone. (k) Differences in transcription factor motif activity comparing erythroid-biased and monocytic biased clones at the earliest sampled time point (day 8).