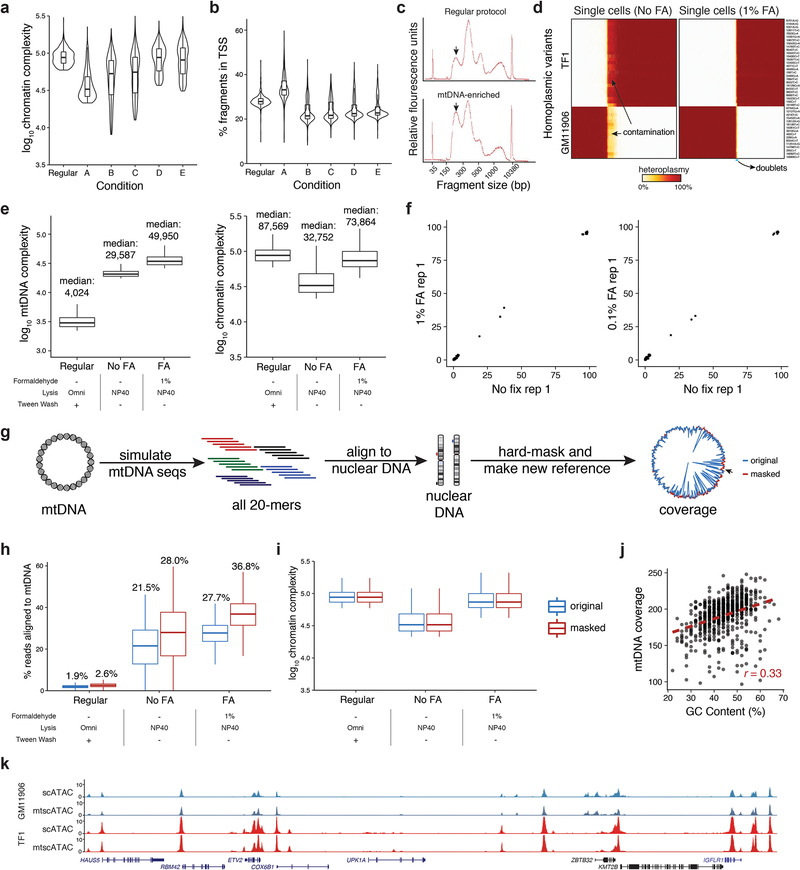
Extended Data Fig. 1: Additional validation of biotechnological and computational basis for single-cell mtDNA genotyping.
(a) Comparison of chromatin library complexity (estimated number of unique fragments) across screened lysis conditions as shown in Fig. 1. (b) The same variable lysis conditions showing the TSS rate per cell. (c) BioAnalyzer traces of mtscATAC-seq library fragment size distribution for regular conditions and mtDNA-enriched conditions. (d) Heteroplasmy heatmap of single cells (columns) for 43 private homoplasmic mutations (rows) in the TF1 or GM11906 cell lines with (left) and without (right) FA treatment. Color bar, heteroplasmy (% allele frequency). (e) Comparison of mtDNA fragment complexity and chromatin complexity between the original/ regular 10x scATAC protocol and modified lysis conditions with and without formaldehyde (FA) treatment. (f) Heteroplasmy of sum of single-cell ATAC-seq libraries with variable FA treatment. (g) Schematic, method, and results of improving mtDNA genome coverage via hard-masking the reference genome (Methods). (h) Comparison of % reads mapping to mtDNA and (i) chromatin complexity with (red) and without (blue) the hard masking. (j) Comparison of average coverage of mtscATAC-seq (y axis) and GC content (x axis) at each 50bp bin (dot) in the mtDNA genome. (k) Accessible chromatin landscapes aggregated from single cells near the ETV2 locus for both cell lines as assayed via regular scATAC-seq and mtscATAC-seq. For boxplots in (a,b,e,h,i), each condition represents the top 1,000 cells (based on chromatin complexity) for one experiment. Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range.