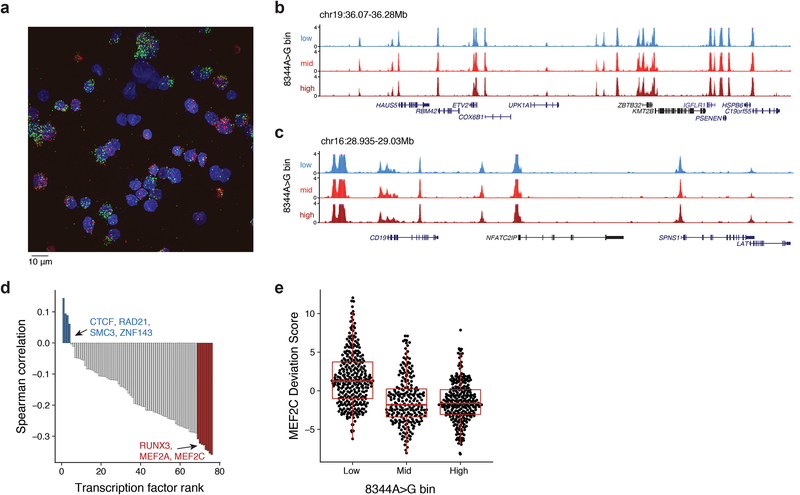
Extended Data Fig. 2: Further inferences in analysis of the GM11906 (MERRF) lymphoblastoid cell line.
(a) Alternative field of view for GM11906 in situ genotyping imaging experiment. Representative image selected from one of seven fields of view for one experiment. Pseudo bulk accessibility track plots are shown for the (b) ETV2 and (c) CD19 loci. Pseudo-bulk groups represent 0–10% (low), 10–60% (mid), and 60–100% (high) m.8344A>G heteroplasmy. (d) Spearman correlation of heteroplasmy against the ChIP-seq deviation scores computed via chromVAR. Each bar is a single transcription factor with selected factors highlighted. (e) Depiction of MEF2C deviation scores from chromVAR for m.8344A>G heteroplasmy bins, corresponding to 0–10% (Low), 10–60% (Mid), and 60–100% (High). Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. Bins contain single cells collected over one experiment where bins correspond to high (>60%; n=273), intermediate (10–60%; n=228), and low (<10%; n=313) heteroplasmy (see Fig. 2c).