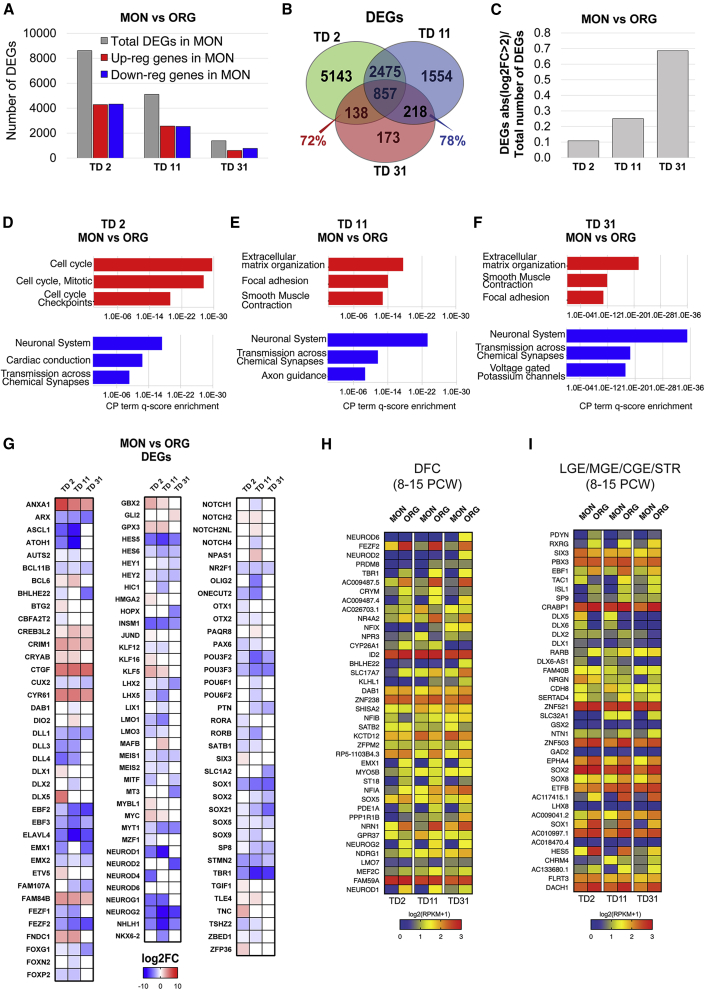
Figure 3.
Comparison of MON and ORG Transcriptional Profiles at Three Different Stages of Neuronal Differentiation (TD2, TD11, and TD31)
(A) Total number of DEGs (gray bar), downregulated DEGs (blue bar), and upregulated DEGs (red bar) in MONs versus ORGs.
(B) Venn diagram of DEGs in MONs versus ORGs at each time point.
(C) Ratio between the number of highly differentially expressed genes (absolute value [log2 fold change > 2]) and the total number of DEGs in MONs versus ORGs along the time course.
(D–F) CP term enrichment for genes upregulated (red) and downregulated (blue) in MONs versus ORGs at each time point, TD2 (D), TD11 (E), and TD31 (F), with the FDR-corrected p value in reverse order on the x axis. For full annotation see Table S3.
(G) Heatmap displaying the log2 fold change values of transcripts from the Neuronal Cell Fate sublist (Table S4A) differentially expressed in MONs versus ORGs.
(H and I) Expression level (log2 (RPKM +1)) in MONs and ORGs of genes that are highly expressed in human dorsolateral prefrontal cortex (DFC) (H) versus basal telencephalon (lateral ganglionic eminence, medial ganglionic eminence, caudal ganglionic eminence, and striatum) or highly expressed in basal telencephalon versus DFC (I).