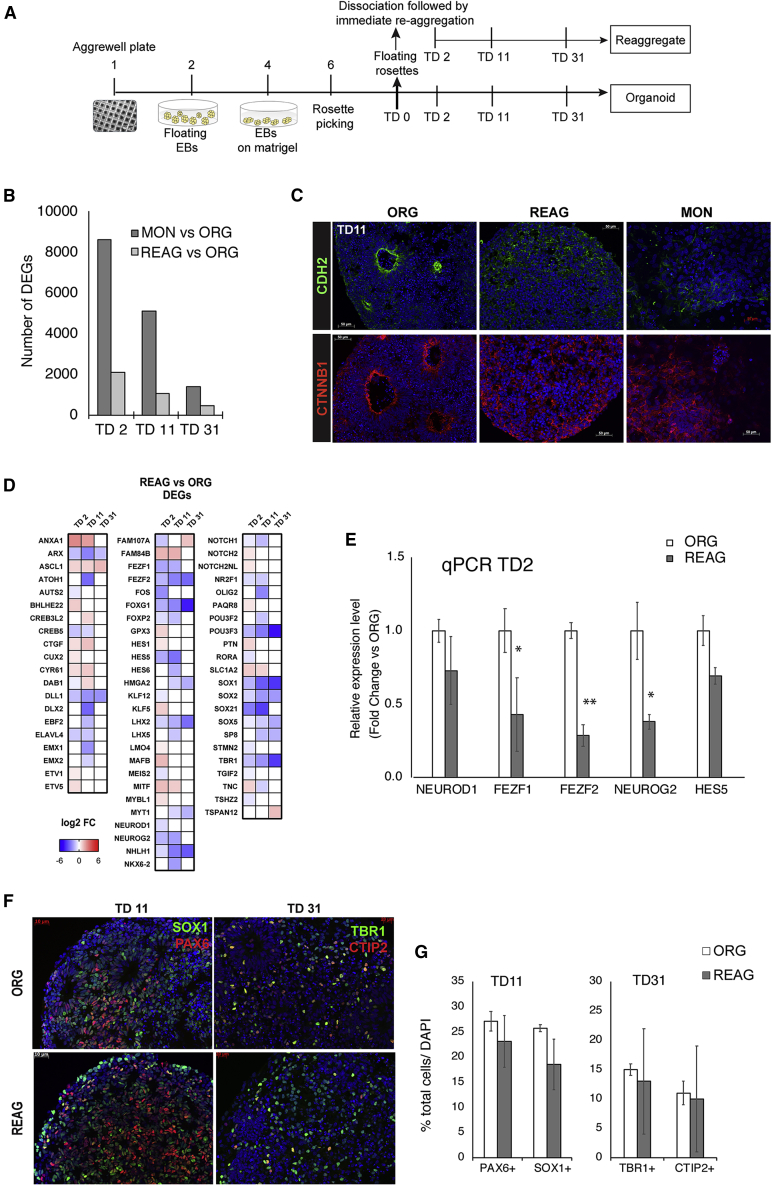
Figure 5.
Dissociation followed by Immediate Reaggregation
(A) Experimental design.
(B) Number of DEGs, at each time point, for the comparisons REAGs versus ORGs and MONs versus ORGs.
(C) Representative images of immunocytochemical staining for N-cadherin (CDH2, green) and β-catenin (CTNNB1, red) at TD11 in ORG, REAGs, and MON preparations (DAPI+ nuclei in blue).
(D and E) Heatmap showing the log2 fold change (D) and bar graph of mRNA expression level by qPCR (E) of DEGs from the Neuronal Cell Fate gene sublist (Table S4A).
(F and G) Representative images of immunocytochemical staining with neuronal progenitor markers (PAX6, SOX1), and the excitatory cortical neuron markers TBR1 and CTIP2 (F) with proportion of different cell types assessed by stereological analysis (G). Results of RNA-seq analysis are from n = 2 biologically different iPSC lines per condition (ORG, REAG) differentiated in parallel.
Immunocytochemical data are expressed as mean ± SEM of n = 3 biologically different iPSC lines per condition (ORG, REAG). Two technical replicates per cell line were analyzed. ∗∗p < 0.01, ∗p < 0.05, MONs versus ORGs analyzed by t test, two tailed. See also Table S5.