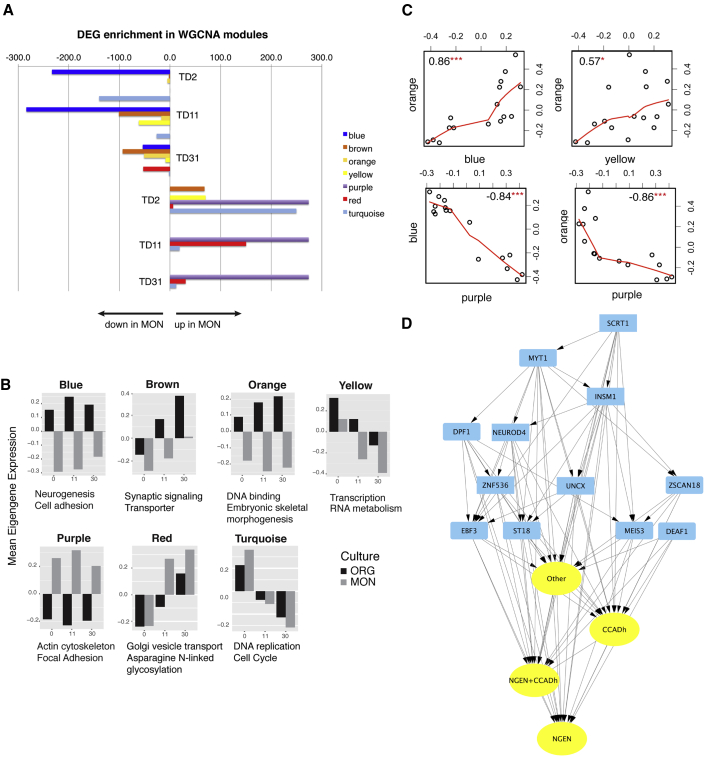
Figure 6.
Characterization of the Seven Transcript Modules Differentially Expressed between MONs and ORGs
(A) Modules' overlap q values (as –log10 [q-value]) with DEGs in MONs versus ORGs at each time point.
(B) Barplots of modules' eigengenes versus time in MONs and ORGs. Reported also are the top scoring functional annotation for each module.
(C) Module to module correlation plots. Represented are the eigengenes as dots and the corresponding correlation coefficients.
(D) Blue module subnetwork, focusing on inferred TFs and associated target genes, as described in Table S7C, after filtering out any edge with an absolute value of the correlation coefficient <0.5. Yellow ovals, cell adhesion (CCADh)-related genes and neurogenesis (NGEN)-related genes, differentially expressed between MONs and ORGs that are TF targets. Blue, upstream TF; arrows, direction of TF-target relationship.