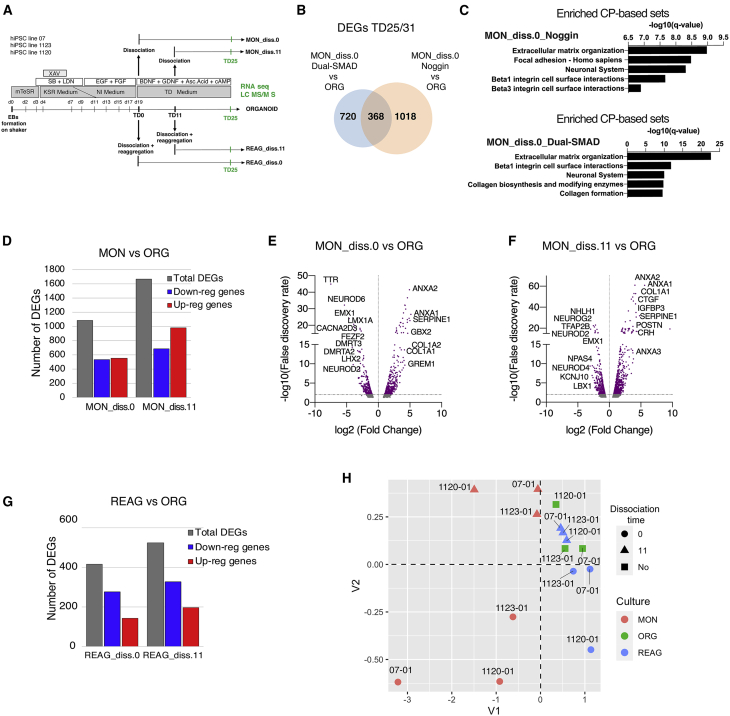
Figure 7.
Consistency of Transcriptomic Changes between MONs and ORGs across Protocols and Times of Dissociation
(A) Experimental design. Cells derived from three iPSC lines were processed for transcriptomic or proteomic analysis. Abbreviations as in the text.
(B) Venn diagram showing overlap between early dissociated MON versus ORG DEGs under Noggin (n = 2 biological different iPSC lines per condition) or Dual-SMAD neuronal induction protocol (n = 3 biologically different iPSC lines per condition).
(C) Top CP-based annotations for the sets of DEGs between early dissociated MONs versus ORGs under Noggin and Dual-SMAD protocols, respectively.
(D) Total number of DEGs (gray bar), downregulated DEGs (blue bar), and upregulated DEGs (red bar) in early and late dissociated MONs versus ORGs at TD25.
(E and F) Volcano plots of MON_diss.0 versus ORG (E) and MON_diss.11 versus ORG (F) DEGs after early and late dissociation. Dots above the horizontal line are statistically significant (FDR < 0.01).
(G) Total number of DEGs (gray bar), downregulated DEGs (blue bar), and upregulated DEGs (red bar) in early and late REAGs versus ORGs at TD25.
(H) Multidimensional scaling plot of RNA-seq data at TD25 for all conditions.