Figure 1.
Efficiency of the Optimized Gastruloid Formation Assay
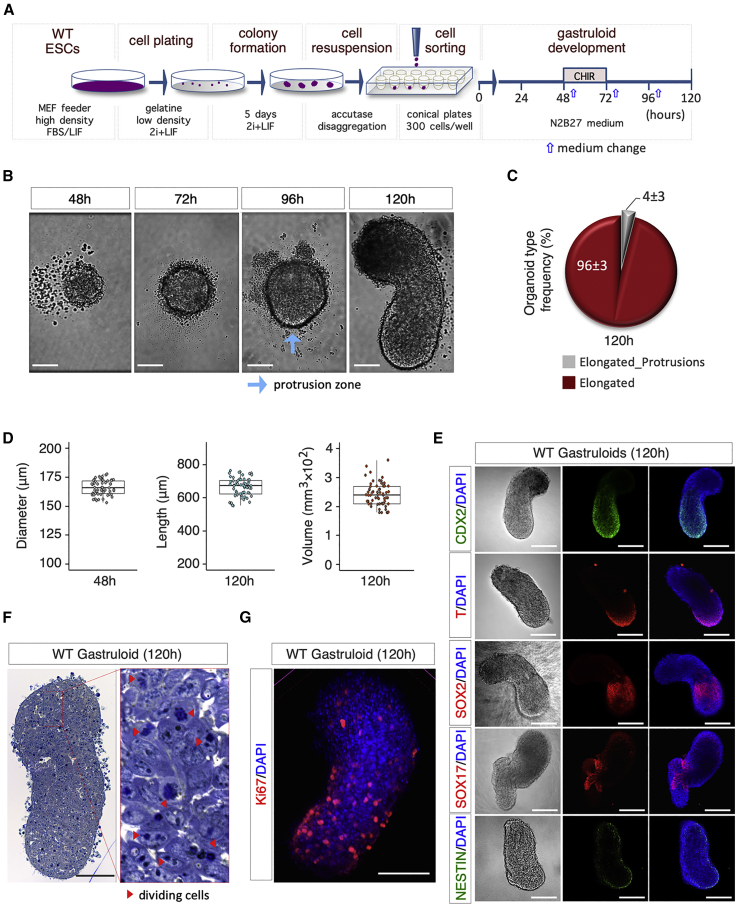
(A) Schematic representation of the experimental design. WT (TBV2) mESCs were plated in 2i + LIF at 250 cells/cm2 on gelatin-coated plates.
(B) Representative bright-field images of aggregate-to-gastruloid transition at the indicated time points after aggregation. Light blue arrow indicates the protrusion zone (bar, 100 μm).
(C) Pie chart quantification of the different organoid phenotypes, i.e., without protrusions with a defined A-P axis (“Elongated”) or with protrusions and a defined A-P axis (“Elongated_Protrusions”). Data are expressed as mean ± SD (n = 3; 180 gastruloids analyzed).
(D) Boxplot diagrams of aggregate diameter distribution at 48 h (left) and gastruloid length (middle) and volume (π·r2·h; right) at 120 h (n = 3; 60 gastruloids/time point).
(E) Representative bright-field (left) and confocal images (middle and right) of gastruloids stained with CDX2, NESTIN (green), T/BRA, SOX2, and SOX17 (red). Nuclei were counterstained with DAPI (bar, 200 μm).
(F) Representative pictures of gastruloid sections stained with toluidine blue; red arrows indicate dividing cells (bar, 100 μm).
(G) Representative confocal image of Ki67 immunostaining of gastruloids. Nuclei were counterstained with DAPI (blue) (bar, 100 μm).
See also Figure S1.