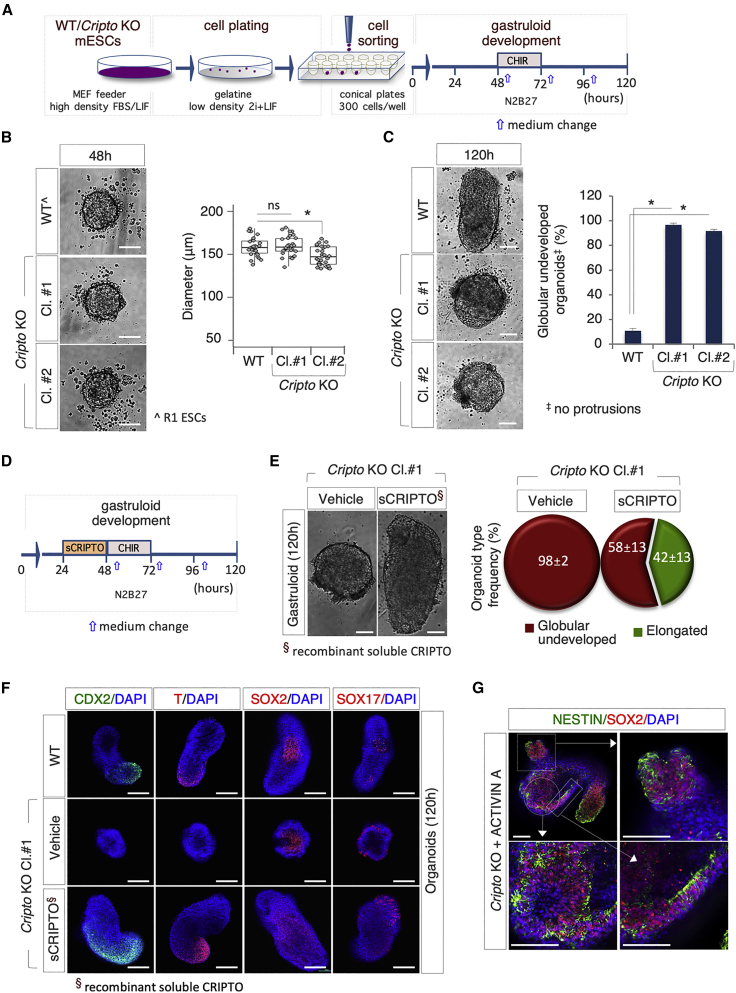
Figure 2.
Cripto Genetic Ablation Impairs Gastruloid Formation
(A) Schematic representation of the experimental design. WT (R1) and Cripto KO mESCs were plated in 2i + LIF at low density on gelatin-coated plates.
(B) Representative bright-field images (left) and diameter distribution (right) of R1 (WT) and Cripto KO clone #1 (Cl.#1) and clone #2 (Cl.#2) mESC-derived aggregates at 48 h (n = 3; 30 gastruloids/condition; ∗p < 0.01; bar, 100 μm).
(C) Representative bright-field images of WT and Cripto KO ESC-derived gastruloids/organoids (left) and quantification (right) of undeveloped WT and Cripto KO organoids at 120 h. Data are expressed as mean ± SD (n = 3; 60 organoids/condition; ∗p < 0.01; bar, 100 μm).
(D) Schematic representation of CRIPTO rescue experiment. Soluble CRIPTO protein (sCRIPTO, 10 μg/mL) was added at 24 h.
(E) Representative bright-field images (left) of Cripto KO ± sCRIPTO-derived gastruloids/organoids. Pie chart quantification of the different organoid type frequencies (right). Data are expressed as mean ± SD (n = 3; 30 gastruloids/condition analyzed; bar, 100 μm).
(F) Representative confocal images of WT and Cripto KO ± sCRIPTO-derived organoids stained with CDX2 (green), T/BRA, SOX2, and SOX17 (red). Nuclei were counterstained with DAPI (blue) (bar, 100 μm).
(G) Representative confocal images of Cripto KO ± ACTIVIN A-derived organoids (120 h AA) with SOX2 (red) and NESTIN (green). Nuclei were counterstained with DAPI (blue) (bar, 100 μm).
See also Figure S2.