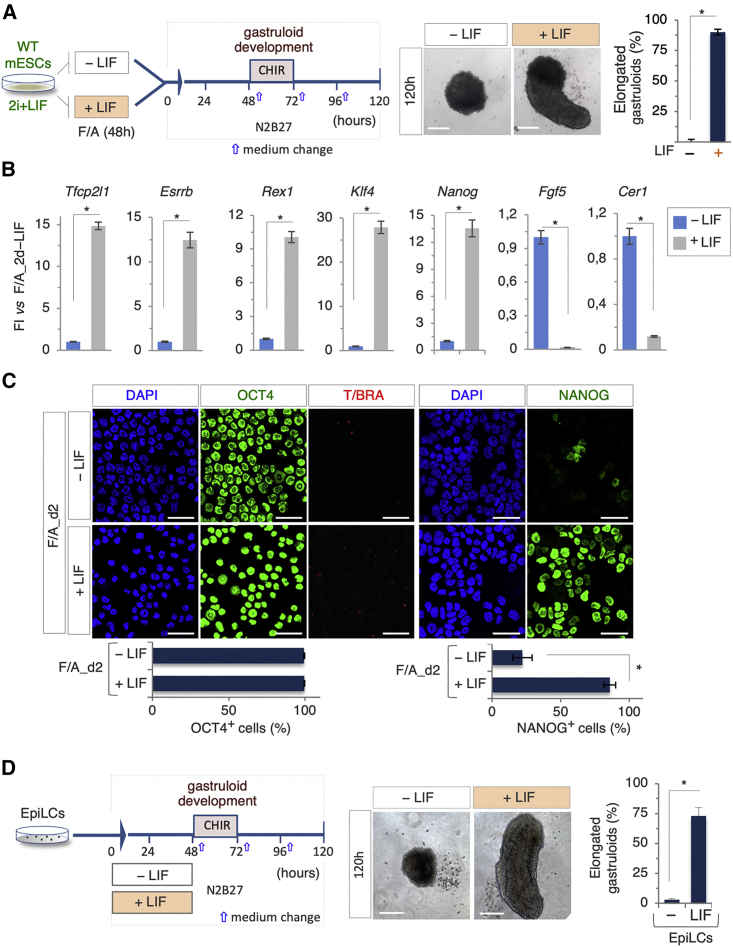
Figure 6.
LIF Dominant Effect on Gastruloid Development
(A) Schematic representation of the experimental design (left). ESCs were treated with FGF/ACTIVIN (F/A) ± LIF for 48 h. Representative bright-field images (middle) of F/A_2d ± LIF-derived organoids and quantification (right) of elongated gastruloids (n = 3; 60 gastruloids/condition; ∗p < 0.01; bar, 100 μm).
(B) Quantitative real-time PCR analysis of pluripotency and differentiation markers in 2d_F/A ± LIF cells. Data represent fold change versus 2d_F/A-LIF; data are normalized to Gapdh and are mean ± SD (n = 3; ∗p < 0.01).
(C) Representative confocal images (top) of OCT4, NANOG (green), and T/BRA (red) staining on cytospun EpiLCs ± LIF. Single-channel images of OCT4 and T/BRA double staining are shown. Nuclei were counterstained with DAPI. Quantification (bottom) of OCT4- and NANOG-positive cells. Data are mean ± SD (bar, 50 μm; n = 3; ∗p < 0.01).
(D) Schematic representation of the experimental design. EpiLC-derived aggregates were treated ± LIF (0–48 h) (left). Representative bright-field images (middle) of EpiLCs ± LIF-derived organoids, and quantification (right) of elongated gastruloids. Data are expressed as mean ± SD (n = 3; 60 gastruloids/condition; ∗p < 0.01; bar, 100 μm).