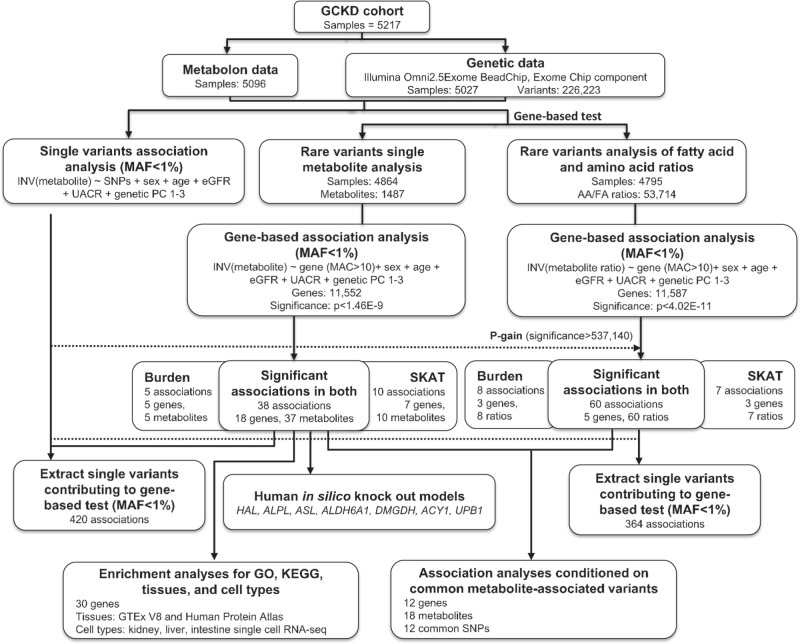
Fig. 1. Rare variant analysis workflow.
The GCKD study enrolled 5,217 patients with moderate CKD. Non-targeted metabolite identification and quantification were conducted from urine samples using the Metabolon HD4 platform. Genotyping was performed with the Illumina Omni2.5Exome Chip. After quality control and data cleaning, genotypes of 226,233 exome chip variants and 1487 metabolites and 53,714 ratios of fatty acids and amino acids were analyzed for 4864 and 4795 patients, respectively. A burden test and the sequence kernel association test (SKAT) were carried out for each gene and each metabolite or metabolite ratio using the seqMeta R package (Methods). Carrier status of variants with minor allele frequency <1% and likely to be functional (splicing, nonsynonymous, stop gain, and stoploss) was evaluated. We used an additive genetic model and adjusted for sex, age, eGFR, UACR, and principal components. Statistical significance was defined using a Bonferroni correction and set at 1.46E−09 (single metabolites) and 4.02E-11 (ratios). Ratio results were further filtered by a p-gain of >537,140 to select ratios that carry information beyond its single metabolites (Methods). The same model was applied to obtain single variant association results for the variants included in the gene-based tests. In silico knockout models to validate findings were generated in a Virtual Metabolic Human. Enrichment analyses of significant genes were carried out using GO terms, KEGG pathways, and gene expression data from tissues and cell types. Conditional analyses were carried out to assess the effect of nearby common metabolite-associated variants on the findings from this study. MAF: minor allele frequency.