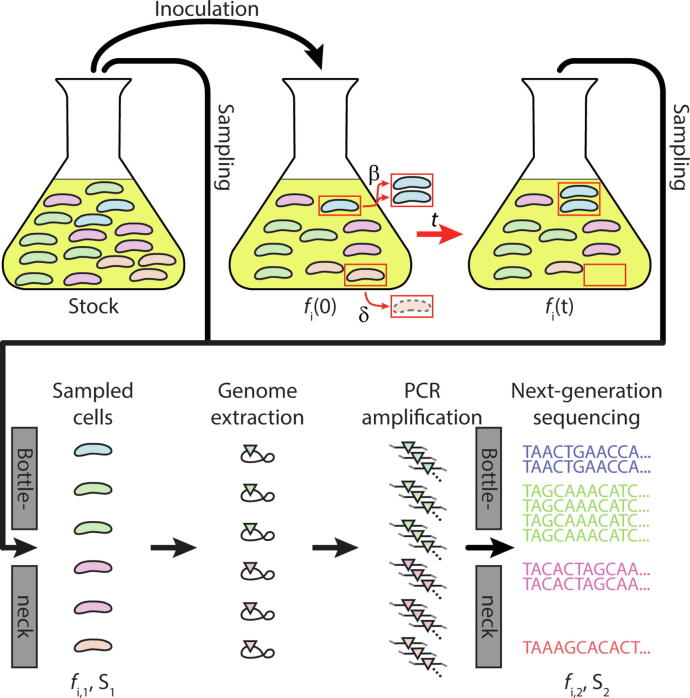
Fig. 2.
The bottlenecks in a typical experimental setup for RESTAMP rate estimates. The schematic lays out a typical experimental setup to determine division and death rates of cells by RESTAMP. An initial population with i = 1,2,…,k = 4 unique sequence tags, indicated by green, purple, beige and blue color, respectively, undergoes a birth–death process (biological bottleneck; indicated by red arrows) for time t with division rate β [min−1] and death rate δ [min−1]. At the beginning of the experiment and at time t, S1 cells are sampled, i.e. pass through a technical bottleneck (small opening in big grey bars). This could for example represent harvesting a set of cells during a time-lapse experiment. This changes the proportion of subpopulation i from fi(t) to fi,1. Following genome extraction (black loops), the genetic tag regions (colored triangles) are amplified by PCR, which we assume is unbiased. Hence, the proportion of the subpopulations do not change and remain fi,1. The amplified tag regions are then sequenced, which constitutes another technical bottleneck (grey bars), where S2 sequence reads are sampled and the proportions are changed from fi,1 to fi,2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)