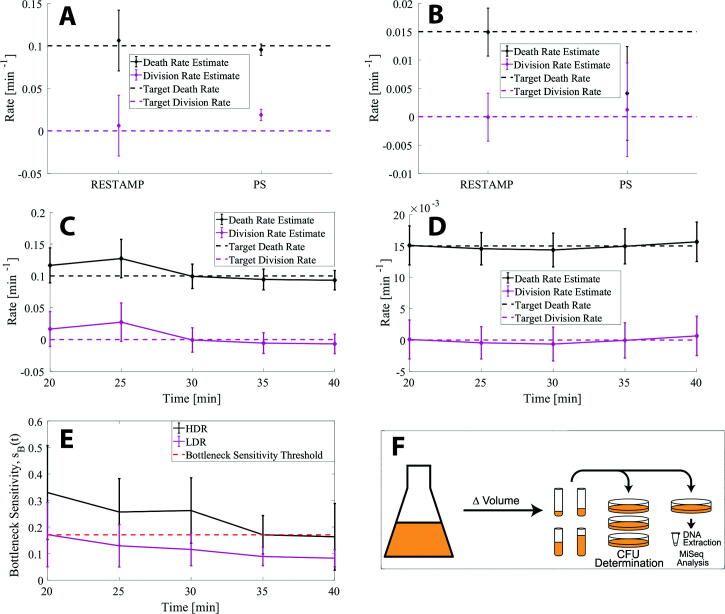
Fig. 4.
The death rates and division rates in an emulated death process are accurately determined by the RESTAMP and PS methods. A pure death process is emulated by sampling different volumes from a starting culture to correspond to different time points in a death process (see 5.1 – Emulating a death process by sampling). (A) The target division rate for a pure death process is 0 (magenta dashed line) and the target death rate was set to 0.1 min−1 (black dashed line). The time points for the RESTAMP experiment were set to t = {20,25,30,35,40} min and 3 repetitions of the experiment were performed. The diamond marker shows the mean estimated rate in a sample size of 15 rates determined for each time point and experiment. The bars show the standard error of the mean. The chosen time points for the plasmid segregation experiment were set to t = {20,40,60,80} min and 3 repetitions of the experiment were performed. (B) Same as (A) except the target death rate was set to 0.015 min−1 and the time points for the plasmid segregation experiments were set to t={20,25,30,35} min. (C) Time resolved division rate and death rate estimates for the high death rate case where the target death rate is 0.1 min−1 (black dashed line) and the target division rate is 0 (magenta dashed line). For each time point, the mean value (diamond marker) and the standard error of the mean are shown. (D) Time resolved division rate and death rate estimates for the high death rate experiment, where the target death rate is 0.015 min−1 (black dashed line) and the target division rate is 0 (magenta dashed line). For each time point, the mean value (diamond marker) and the standard error of the mean are shown. (E) The bottleneck sensitivity was calculated using equation (8) for the individual high death rate (HDR – black solid line) and the low death rate experiments (LDR – magenta solid line) (Figure Supplementary Fig. 4) where the plot shows the mean bottleneck sensitivity ± S.D. A threshold value of sB(t) = 0.17 for the bottleneck sensitivity was set to correspond to robust rate estimates (red dashed line) at t = 35 min. (F) A simple schematic of the experiment. Different size volumes (Δvt) are sampled from an Erlenmeyer flask which contains either a population of cells with a sequence tag (STAMP) or a population of cells with an identifiable plasmid (PS) in LB media. The size of the volumes was determined so as to emulate a pure death process (see 5.1 – Emulating a death process by sampling). Colony forming units (CFUs) were determined by serial dilution for the largest volume (corresponding to t = 0) and extrapolated to the pre-determined time points in (C-D). For the RESTAMP method, the genomes were extracted and tag frequencies were determined by next-generation sequencing. For the PS method the fraction of cells carrying the conditionally replicative plasmid were determined by selective plating. The experiments were repeated biologically independently three times. Rate estimates for all trials at each time point and experimental CFU and NB values are available for download on SourceForge (see 6 – Code). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)