Figure 2 |. TLR4 controls the expression of epithelial Duox2 and Nox1.
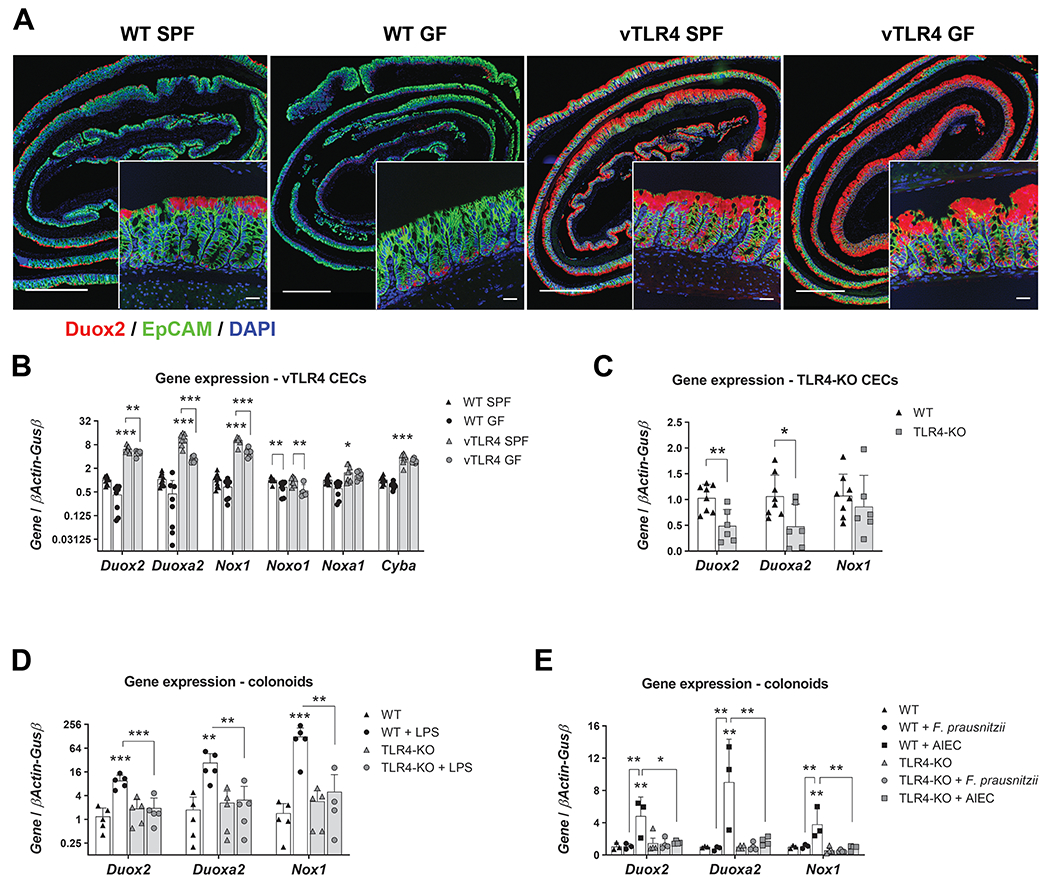
(A) Representative micrographs show Duox2 transcripts (red) counterstained with EpCAM (green) and DAPI (blue). Micrograph scale bar = 1 mm; inset scale bar = 25 μm. (B) Freshly isolated CECs were analyzed by qRT-PCR for the expression of selected transcripts. Villin-TLR4 SPF CECs showed increased expression of Duox2, Duoxa2, Nox1, Cyba (***P<0.001), and Noxa1 (*P<0.05) when compared to WT SPF CECs (n=8-10 mice). Data were analyzed by two-way ANOVA followed by Sidak’s post-hoc test for each gene. “Microbiota” (SPF vs GF) was identified as a significant source of variation in the expression of Duox2, Duoxa2, Nox1, Noxo1 (all P<0.001), Noxa1, and Cyba (both P<0.05); n=6-10 mice. (C) Gene expression in freshly isolated CECs of TLR4-KO and WT mice. Duox2 (**P<0.01) and Duoxa2 (*P<0.05) were significantly downregulated in TLR4-KO CECs (n=6-8 mice). Data were analyzed by unpaired t-test for each gene. (D) Cultured WT and TLR4-KO colonoids were stimulated with LPS for 24 hours and their gene expression was determined by qPCR. Duox2, Nox1 (***P<0.001), and Duoxa2 (**P<0.01) transcripts were significantly upregulated by LPS in WT colonoids (n=5 cultures). Data were analyzed by two-way ANOVA followed by Sidak’s post-hoc test for each gene. (E) Cultured WT and TLR4-KO colonoids were stimulated with heat killed F. prausnitzii or AIEC for 24 hours and their gene expression was determined by qPCR. Duox2, Duoxa2, and Nox1 (**P<0.01) transcripts were significantly upregulated by AIEC in WT colonoids (n=3-4 cultures). Data were analyzed by two-way ANOVA followed by Sidak’s post-hoc test for each gene.
